Abstract
Apium graveolens L. is a worldwide popular vegetable with pharmacological efficacy. The complete chloroplast genome of A. graveolens was determined in this study. The total genome size was 152,050 bp in length, containing a pair of inverted repeats (IRs) of 24,743 bp, which were separated by large single copy (LSC) and small single copy (SSC) of 85,441 bp and 17,123 bp, respectively. The GC content is 37.64%. A total of 129 genes were predicted including 84 protein-coding genes, eight rRNA genes and 37 tRNA genes. Phylogenetic tree analysis of 30 species in the family Apiaceae indicated that A. graveolens was closer to Anethum, Foeniculum and Petroselinum.
Apium graveolens is a plant species in the family Apiaceae commonly known as celery (var. dulce), originating from Mediterranean of southern Europe and the swamp of Egypt and Sweden (Wang et al. Citation2011; Bankevich et al. Citation2012; Li et al. Citation2016). Celery is rich in a variety of nutrients like vitamins and minerals, which make it an increasing popular vegetable to consumers, and contained many secondary metabolites with pharmacological efficacies, such as apiin, flavonoids, carotenoids and volatile oils, which make it an important medicine for curing inflammatory, rheumatism and arthritis (Li et al. Citation2016; Mezeyova et al. Citation2018). Chloroplast genome was an important part of plant whole genome, the gene on the genome coding many proteins is necessary for the process of photosynthesis, and the genome sequence is a great molecular resource for plant taxonomy, genetic diversity and phylogenetic system researches. Therefore, we reported complete sequence of the chloroplast genome of celery with hope to promote these researches based on the celery chloroplast genome in this study.
Sample of A. graveolens (accession no. JA-cy01) was stored in the college of agronomy of Jiangxi Agricultural University (28°45′27″N, 115°50′20″E), Nanchang, China. Genomic DNA was extracted from fresh leaves and subjected to construct a genomic library and sequenced by HiSeq X Ten (BioMarker, Beijing, China). About 1 Gb of sequence data with ∼2× coverage to its nuclear genome was obtained after sequencing and base quality control, 0.5 Gb of clean pair-end reads was randomly extracted using Seqtk. The celery chloroplast genome was assembled by using SPAdes (v 3.12.0)(Bankevich et al. Citation2012), BlastN (v2.7.1), and Gapcloser (v1.12-r6). Firstly, these reads were assembled with using the Plasmidspades.py in SPAdes. Secondly, contigs representing the chloroplast genome were retrieved, ordered and incorporated into a single draft sequence by comparison with the chloroplast genome of Daucus carota (NC_008325.1) using BlastN. Thirdly, the gaps in the chloroplast single draft sequence of were closed by using GapCloser. Finally, the complete genome sequence was annotated using GeSeq(Tillich et al. Citation2017) and manually corrected by visual inspection using IGV(Robinson et al. Citation2011).
A typical chloroplast genome consists of four distinct regions, a large and a small single copy region (LSC and SSC, respectively) separated by two inverted repeat regions (IRa and IRb). The complete chloroplast genome of A. graveolens (accession no. MK036045) is 152,050 bp in length with 37.64% GC contents and exhibits a typical quadripartite structure, consisting of a pair of IRs (24,743 bp) separated by the LSC (85,441 bp) and SSC (17,123 bp) regions. There is a total of 129 genes, including 86 protein-coding genes, eight rRNA genes and 37 tRNA genes; six of the protein-coding genes, six of the tRNA genes and four rRNA genes are duplicated within the IRs.
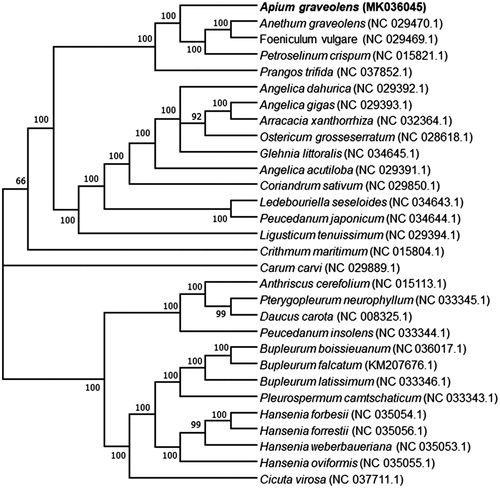
To determine the phylogenetic position of A. graveolens, a phylogenetic analysis was carried out among 29 complete chloroplast genomes of Apiaceae that are directly downloaded from the organelle genome database in NCBI. The phylogenetic tree was constructed by Maximum Likelihood method using the programme MAFFT v7.407(Nakamura et al. Citation2018) and MEGA v10.0.4(Kumar et al. Citation2018). The results showed that A. graveolens was closer to Anethum, Foeniculum and Petroselinum, but has remote phylogenetic relationship with these genera, Hansenia, Cicuta, Pleurospermum, Bupleurum, Daucus, Pterygopleurum and Anthriscus in the family Apiaceae ().
Figure 1. Phylogenetic tree showing relationship between Apium graveolens and other 29 species in the family Apiaceae. Phylogenetic tree was constructed based on the complete chloroplast genomes using maximum likelihood (ML) with 1000 bootstrap replicates. Numbers in each the node indicated the bootstrap support values.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol Comput Biol. 19:455–477.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Li MY, Feng W, Qian J, Wang GL, Chang T, Xiong AS. 2016. Validation and comparison of reference genes for qPCR normalization of celery (Apium graveolens) at different development stages. Front Plant Sci. 7:313.
- Mezeyova I, Hegedűsová A, Mezey J, Šlosár M, Farkaš J. 2018. Evaluation of quantitative and qualitative characteristics of selected celery (Apium graveolens var. Dulce) varieties in the context of juices production. Potravinarstvo. 12:173–179.
- Nakamura T, Yamada KD, Tomii K, Katoh K. 2018. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics. 34:2490–2492.
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol. 29:24–26.
- Tillich M, Lehwark P, Pellizzer T, Ulbrichtjones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:(W1):W6-W11.
- Wang S, Yang W, Shen H. 2011. Genetic diversity in Apium graveolens and related species revealed by SRAP and SSR markers. Sci Hortic. 129:1–8.
