Abstract
The complete mitochondrial genome of the saw-jawed monocle bream, Scolopsis ciliata was determined by the next generation sequencing (NGS) technique. The circular mitogenome of S. ciliata (16,733 bp) encoded the canonical 37 genes including 13 proteins, 22 tRNA genes, 2 rRNAs (12S rRNA and 16S rRNA), and two non-coding regions; the origin of light strand replication (OL) and the putative control region (D-Loop). The gene arrangement of the S. ciliata mitogenome was identical to its relative, Scolopsis vosmeri. Phylogenetic analysis based on the full mitochondrial genome sequences showed that S. ciliata is most closely related to S. vosmeri with 83% nucleotide sequence identity. The mitogenome information of S. ciliata would be the useful information to understand the evolutional relationship of fishes in genus Scolopsis.
Approximately 17 species are currently known in the genus Scolopsis and most of which inhabit widly in Indo-West Pacific region (Russell Citation1990). The Scolopsis ciliata is one of economically important marine fishes in Indonesia, which found on the sandy fringe of coastal and lagoon reefs (Kuiter and Tonozuka Citation2001; Allen and Erdmann Citation2012). Currently, phylogenetic relationship of fishes in genus Scolopsis is still not clearly eastablished (Chen and Zheng Citation1987; Russell Citation1990) and only one mitochondrial genome sequence in the genus, Scolopsis vosmer is currently reported (Wu et al. Citation2017). In this study, we report a mitogenome sequence of S. ciliate, which would be the useful information to understand the evolutional relationship of fishes in genus Scloposis.
The next-generation sequencing (NGS) platform was adopted to read the complete mitochondrial genome sequence of C. ciliata. The specimen was collected from the coastal water in Malang, East Java, Indonesia (8°26'05,65"S 112°40'55,31"E) and deposited at the Ichtyology Laboratorium, Universitas Airlangga, Indonesia. Species of the specimen was identified by both its morphological characteristics and DNA sequence identity in COI region to the database (GenBank Accession number: KY362945). Purified mitochondrial DNA by the commercial kit (Abcam, Cambridge, UK) was further fragmented into to the size for library preparation (∼350 bp) by Covaris M220 Focused-Ultrasonicator (Covaris Inc., Woburn, MA). A library was constructed by TruSeq® RNA library preparation kit V2 (Illumina, San Diego, CA) and its quality and the quantity was analysed by 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). High-throughput sequencing was performed by Illumina MiSeq system (2 × 300 bp).
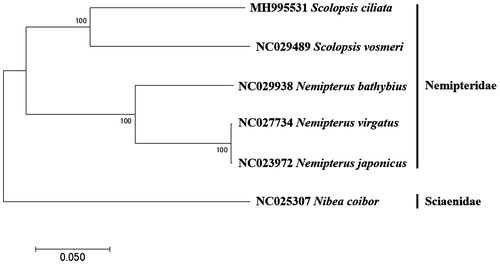
The complete mitochondrial genome of S. ciliata was circular and 16,733 bp in length (GenBank Number: MH995531). It contained the typical 37 genes coding for two ribosomal RNAs (12S rRNA and 16S rRNA), 22 tRNAs, 13 polypeptides, and two non-coding regions; the origin of light strand replication (OL) and the putative control region (D-Loop). OL was located between tRNA-Asn and tRNA-Cys at WANCY cluster, while D-Loop (986 bp) was identified between tRNA-Pro and tRNA-Phe. Besides COX1(GTG) and ATP6 (ATC), 11 protein-coding genes begin with typical ATG. Incomplete stop codons were shown in ND2, COX2, COX3, ND3, ND4, and CYTb genes. The length of tRNAs ranged between 69 and 76 bp, which were located as three conserved tRNA cluster (IQM, WANCY, and HSL)(Satoh et al. Citation2016). Except for tRNA-Ser, 21 tRNAs were predicted to form the typical clover-leaf secondary structures according to ARWEN (Laslett and Canbäck Citation2008) which has the typical clover-leaf secondary structures except for tRNA-Ser. Based on the phylogenetic analysis of the mitogenomes by MEGA7 version 7 software (Kumar et al. Citation2016), S. ciliata was most closely related to S. vosmeri (GenBank No. NC029489) with 83% identity forming a genus Scolopsis cluster, which is distinct from fishes in genus Nemipterus (). The mitogenome information of S. ciliata would help us understand the evolutional relationship in genus Scolopsis.
Disclosure statement
The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Additional information
Funding
References
- Allen GR, Erdmann MV. 2012. Reef fishes of the East Indies. Perth, Australia: Tropical Reef Research.
- Chen Q, Zheng B. 1987. Systematic synopsis of Chinese fishes. Beijing: Science Press; p. 1–2.
- Kuiter R, Tonozuka T. 2001. Indonesian reef fishes (part 1 dan 2). Melbourne, Australia: Zoonetic.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Russell BC. 1990. Nemipterid fishes of the world. Vol. 12. Rome, Italy: FAO Fisheries Synopsis; p. I.
- Satoh TP, Miya M, Mabuchi K, Nishida M. 2016. Structure and variation of the mitochondrial genome of fishes. BMC Genom. 17:719–738.
- Wu R, Liu J, Niu S, Zheng J. 2017. The complete mitochondrial genome of Scolopsis vosmeri and phylogenetic relationship of genus Scolopsis. Mitochondrial DNA A DNA Mapp Seq Anal. 28:81–82.