Abstract
The complete mitochondrial genome sequence of Dory snapper, Lutjanus fulviflamma, was determined by high-throughput sequencing (HTS) technique. The circular mitogenome of L. fulviflamma was 16,512 bp in length encoding 37 genes (13 proteins, 22 tRNAs, 2 ribosomal RNAs) and 2 conserved noncoding elements; the control region (D-Loop) and the origin of light strand synthesis (OL). Among the protein-coding genes, unusual start codons were identified in ND2, COX1 and ND3 genes and incomplete stop codons were identified in seven genes. Phylogenetic analysis with the currently known mitogenomes in genus Lutjanus revealed that L. fulviflamma was most closely related to Lutjanus russellii with 91% sequence identity.
Fishes in genus Lutjanus are usually found in tropical and subtropical reefs or mangrove forests in the Atlantic, Indian and Pacific Oceans. Among 73 currently known species in the genus, Dory snapper, Lutjanus fulviflamma is commercially and ecologically important species in western Pacific and Indian oceans (Iwatsuki et al. Citation1993; Salini et al. Citation2006). As shown in its relatives, L. fulviflamma shows a seasonal migrating pattern in and out of the coral reefs for its reproduction (Grol et al. Citation2008; Nagelkerken Citation2009). In order to know the genetic population of L. fulviflamma from a variety of its relatives, which share the habitat, molecular identification strategy would be more effective than morphological analysis. We, here, report the full mitochondrial genome sequence of L. fulviflamma, which was determined by high-throughput sequencing (HTS) technology.
The specimen was collected from the coastal water in Muncar Banyuwangi, East Java, Indonesia (8°12′07,52ʺS 114°23′07,18ʺE) and stored at Universitas Airlangga, Indonesia. Identification of the specimen was made by both the morphological characteristics and the sequence identity in COI barcode region to the database (GenBank Accession number: MG002617). Mitochondrial DNA of L. fulviflamma was extracted by the mitochondrial DNA isolation kit (Abcam, UK) and further fragmented into smaller sizes (∼350 bp) by Covaris M220 Focused-ultrasonicator (Covaris Inc., Woburn, MA). A library for the sequencing was constructed by TruSeq® RNA library preparation kit V2 (Illumina, CA) and its quality and the quantity was analyzed by 2100 Bioanalyzer (Agilent Technologies, USA). DNA sequencing was performed by MiSeq sequencer (Illumina, CA).
The complete mitochondrial genome of L. fulviflamma (GenBank Number: MH995530) was 16,512 bp in length, which consisted of 13 protein-coding genes, 22 tRNAs, two ribosomal RNAs (12S and 16S), and two non-coding elements; origin of L strand replication (OL) and putative control region (D-Loop). Except for tRNA-Ser(GCT), all the tRNAs were predicted to form a three-armed clover structures by ARWEN software (Laslett and Canbäck Citation2008; Satoh et al. Citation2016). Ten protein-coding genes begin with typical ATG start codons and incomplete stop codons were identified in ND2, COX2, ATP6, COX3, ND3, ND4, and Cyt b genes. Two ribosomal RNAs (12S rRNA and 16S rRNA) were located between tRNA-Phe(GAA) and tRNA-Leu(TAA). The non-coding region (OL) and D-Loop was identified between tRNA-Asn and tRNA-Cys at WANCY cluster and between tRNA-Pro and tRNA-Phe, respectively, as shown in the mitogenome of its relative, Lutjanus vitta (Andriyono et al. Citation2018).
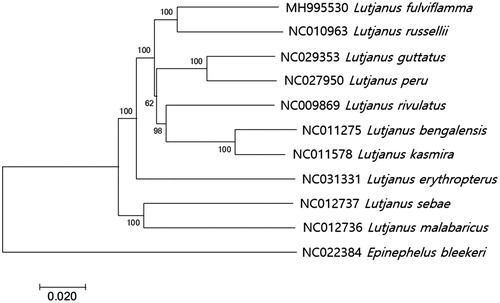
The phylogenetic analysis of the currently known mitogenome sequences of fishes in Lutjanidae showed that L. fulviflamma was most closely related to Lutjanus russellii (GenBank Number: NC010963) with 91% nucleotide sequence identity followed by Lutjanus guttatus (GenBank Number: MH675887, 90% identity) (). This mitogenome information of L. fulviflamma would provide basic information for its scientific management and conservation in Indonesia.
Figure 1. Phylogenetic tree of Lutjanus fulviflamma within Lutjanidae. Phylogenetic tree of Lutjanus fulviflamma complete genome was constructed by MEGA7 software with Minimum Evolution (ME) algorithm with 1000 bootstrap replications. GenBank Accession numbers were shown followed by each scientific name and furthermore Epinephelus bleekeri (NC022384) as an outgroup.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Andriyono S, Alam J, Kwak DH, Kim H-W. 2018. Complete mitochondrial genome of brownstripe red snapper, Lutjanus vitta (Perciformes: Lutjanidae). Mitochondrial DNA Part B. 3:1129–1130.
- Grol MG, Dorenbosch M, Kokkelmans EM, Nagelkerken I. 2008. Mangroves and seagrass beds do not enhance growth of early juveniles of a coral reef fish. Mar Ecol Prog Ser. 366:137–146.
- Iwatsuki Y, Akazaki M, Yoshino T. 1993. Validity of a lutjanid fish, Lutjanus ophuysenii (Bleeker) with a related species, L. vitta (Quoy et Gaimard.). JPN J Ichthyol. 40:47–59.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Nagelkerken I. 2009. Evaluation of nursery function of mangroves and seagrass beds for tropical decapods and reef fishes: patterns and underlying mechanisms, Ecological connectivity among tropical coastal ecosystems. Springer: Heidelberg; p. 357–399.
- Salini J, Ovenden J, Street R, Pendrey R. 2006. Genetic population structure of red snappers (Lutjanus malabaricus Bloch & Schneider, 1801 and Lutjanus erythropterus Bloch, 1790) in central and eastern Indonesia and northern Australia. J Fish Biol. 68:217–234.
- Satoh TP, Miya M, Mabuchi K, Nishida M. 2016. Structure and variation of the mitochondrial genome of fishes. Bmc Genomics. 17:719–738.
