Abstract
The complete mitochondrial genome was sequenced from the marine teleost fish Black Sea Bass, Centropristis striata. The genome sequence is 16,986 bp in size and has a base composition of A (26.65%), T (26.44%), C (28.63%), and G (18.28%). The phylogenetic analysis showed that C. striata belongs to Serranidae family and is most closely related to Hypoplectrus. The complete mitochondrial genome sequences provided here was the first one in Centropristis, which would be useful for further understanding the evolution and conservation genetics of C. striata.
The black sea bass (Centropristis striata, Serranidae, Perciformes) is a kind of high-valued marine teleost fish that support important commercial and recreational fisheries along the Atlantic coast of the USA and the east of China (Morin et al. Citation2015). Although its biological characteristics are very near to Epinephelus, in which the complete mitochondrial genome is reported in several species (Du et al. Citation2018; Kim et al. Citation2016a), none of Centropristis species have complete mitochondrial genome sequenced. In this study, we first characterized the complete mitochondrial genome sequences of C. striata to contribute to further physiological, molecular and phylogenetical studies of this fish.
The C. striata in this study were artificially bred in fisheries research institute of Zhoushan City in Zhujiajian aquaculture base, Zhejiang Province of China. The parent fish were from the Atlantic Ocean (31°05′33.8ʺN 75°15′41.6ʺW). Samples stored in a refrigerator of 80 °C with accession number 20180711EC03.Genomic DNA was extracted by using the Qiagen DNA extract kit (Qiagen DNeasy, Hilden, Germany) following the instructions of the manufacturer and then subjected to build up the genomic library and pair-end sequencing (2 × 150 bp) by MiSeq (Illumina, San Diego, CA). Approximately 23 Mb of raw data and 21.9 Mb clean data were obtained, and de novo assembled by the SOAPdenovo software (Version 2.04, http://soap.genomics.org.cn/soapdenovo.html). DOGMA software was used for annotation of protein-coding genes (PCGs) in the mitochondrial genome (Wyman et al. Citation2004), and manually inspected to predict PCGs, transfer RNA (tRNA) genes, and ribosomal RNA (rRNA) genes.
The complete mitochondrial genome of C. striata was 16,986 bp in size, with a genome size similar to Epinephelus. The accurate annotated mitochondrial genome sequence was submitted to GenBank with the accession number MH645336. The complete mitogenome has 13 PCGs, 22 tRNA genes, and 2 rRNA genes. Genes encoding on the genome are similar among all Perciformes (Ayala et al. Citation2017; Kim et al. Citation2016b). The total base composition of the mitogenome is A (26.65%), T (26.44%), C (28.63%), and G (18.28%). Which shows AT bias, with the AT content of 53.1%. The percentage of GC (46.9%) was a little lower than AT rate, which was generally in accordance with other teleost mitochondrial genomes (Sui et al. Citation2017).
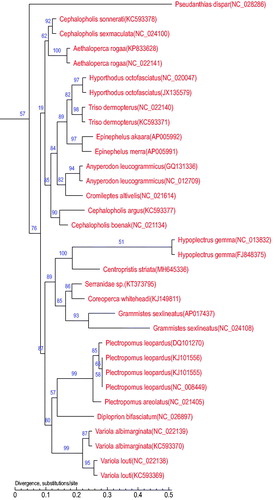
To further validate the new sequences and the phylogenetic position of C. striata, we used the mitochondrial genomes of 32 species which from 16 different genera in Serranidae to construct the phylogenetic tree (). we constructed the phylogenetic tree using maximum likelihood (ML) method in the MEGA package (Kumar et al. Citation2016) with bootstrap values of 1000 replicates. The tree showed that C. striata was most closely related to Hypoplectrus species, they both belong to Serraninae subfamily. Above all, this work is the first to sequence the mitochondrial genome in Centropristis, which will provide important information for its evolutionary and genomic studies.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Ayala L, Becerra J, Boo GH, Calderon D, DiMarco A, Garcia A, Gonzales R, Hughey JR, Jimenez GA, Jimenez TA, et al. 2017. The complete mitochondrial genome of Xiphister atropurpureus (Perciformes: Stichaeidae). Mitochondrial DNA Part B. 2:161–162.
- Du X, Gong L, Chen W, Liu L, Lü Z. 2018. The complete mitochondrial genome of Epinephelus Chlorostigma (Serranidae; Epinephelus) with phylogenetic consideration of Epinephelus. Mitochondrial DNA Part B. 3:209–210.
- Kim YK, Lee YD, Oh HS, Han SH. 2016a. The complete mitochondrial genome and phylogenetic position of the Endangered red-spotted grouper Epinephelus akaara (Perciformes, Serranidae) collected in South Korea. Mitochondrial DNA Part B. 1:927–928.
- Kim YK, Song YS, Kim IH, Kang CB, Kim WB, Kim SY. 2016b. Complete mitochondrial genome of Lycodes tanakae (Perciformes: Zoarcidae). Mitochondrial DNA Part B. 1:947–948.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Morin SJ, Decatur WA, Breton TS, Marquis TJ, Hayes MK, Berlinsky DL, Sower SA. 2015. Identification and expression of GnRH2 and GnRH3 in the black sea bass (Centropristis striata), a hermaphroditic teleost. Fish Physiol Biochem. 41:383–395.
- Sui S, Yang Y, Fang Z, Wang J, Wang J, Fu YQ, Hou Y, Xu B, Yu J. 2017. Complete mitochondrial genome and phylogenetic analysis of Ixodes persulcatus (taiga tick). Mitochondrial DNA Part B. 2:3–4.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.