Abstract
We performed high-throughput sequencing on the complete mitogenomes of two raptors, white-tailed eagle (Haliaeetus albicilla) and oriental honey buzzard (Pernis ptilorhynchus). This is the first time that the complete mitogenomes from Accipitridae subfamies, Haliaeetinae, and Perninae have been sequenced. Respectively, H. albicilla and P. ptilorhynchus mitogenomes were 17,719 and 17,927 bp base pairs long. The ND3 gene in both raptors possessed an insertion mutation, whereas only P. ptilorhynchus ND6 had two mutations. Our results benefit future raptor research through expanding the mitogenome database.
The white-tailed eagle (Haliaeetus albicilla Linnaeus, 1758; Haliaeetinae, Accipitridae) is one of the largest sea eagles widely distributed across Eurasia. Northern populations breed in northeast Russia and northeast China but migrate to southeast China, Korea, and Japan to winter (Ferguson-Lees and Christie Citation2001; Brazil Citation2009). The oriental honey buzzard (Pernis ptilorhynchus Temminck, 1821; Perninae, Accipitridae) breeds in the Russian far east, north Mongolia, northeast China, Korea, and Japan, but then migrate to Southeast Asia to winter (Ferguson-Lees and Christie Citation2001; Brazil Citation2009). Although listed as least concern in the International Union for Conservation of Nature (IUCN) Red List (IUCN Citation2018a, Citation2018b). In South Korea’s Ministry of Environment has classified H. albicilla and P. ptilorhynchus as endangered species category I and II, respectively (PNIBR Citation2017). In this study, we used Illumina HiSeq to sequence the complete mitochondrial genomes of both raptors. With these sequences and publicly available GenBank data, we confirmed their phylogenetic relationships among Accipotridae Samples from H. albicilla (IN1796) and P. ptilorhynchus (IN1589) were collected in Seosan-si Chungnam, South Korea and deposited in the National Institute of Biological Resources at Inchoen, South Korea. Genomic DNA was extracted from blood using a DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA) in accordance with the manufacturer protocol. Macrogen (Seoul, South Korea) performed whole genome resequencing with the Illumina HiSeq 2500 platform. DOGMA (Wyman et al. Citation2004) and ARWEN (Laslett and Canback Citation2008) were used for annotation.
Mitogenome length and AT content for H. albicilla are 17,719 bp and 54.25%, respectively (GenBank accession: MK043028); for P. ptilorhynchus, 17,927 bp and 53.14% (MK043029). The structures of both mitogenomes are typical of Accipitridae (Mindell et al. Citation1998a) comprising 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes, and two non-coding control regions. Similar to several avian species (Mindell et al. Citation1998b; Jeon et al. Citation2018) an insertion mutation is present in the mitochondrial ND3 gene of both raptors. In P. ptilorhynchus specifically, the ND6 gene exhibits a 2 bp deletion and a 1 bp insertion.
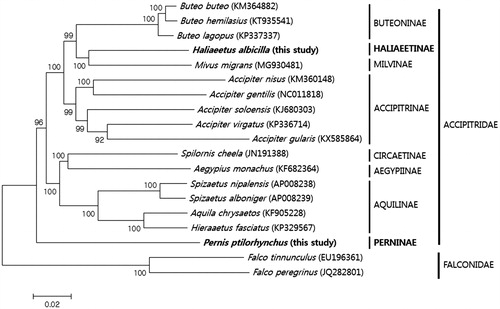
We constructed a neighbour-joining tree in MEGA version 6.0 (Arizona State University, Tempe, AZ), using the 13 protein-coding genes and two ribosomal RNA genes sequenced here, along with 17 Accipitridae mitogenomes, downloaded from GenBank (). Phylogenetic analysis strongly confirmed the monophyly of Haliaeetinae and Perninae. The former subfamily clustered closely with Milvinae and Buteoninae, whereas Perninae was distant. Our findings are consistent with previous results (Lerner and Mindell Citation2005). In conclusion, this study provides two new additions to the avian mitogenome database, thus enriching an important resource in evolutionary and ecological studies.
Figure 1. The consensus neighbor-joining phylogenetic tree of eight Accipitridae subfamilies. Accipitridae mitogenome sequences were from this study (in bold) and GenBank; two Falco mitogenomes, also from GenBank, were used as outgroups. Number on the nodes is bootstrap values, estimated for concatenated sequences of 13 protein-coding genes and two ribosomal RNA genes.
Disclosure statement
The authors report no conflicts of interest and are alone responsible for the content and writing of the article.
Additional information
Funding
References
- Brazil M. 2009. Birds of East Asia. London: Christopher Helm Ltd; p.120–125.
- Ferguson-Lees J, Christie DA. 2001. Raptors of the world. London: Christopher Helm Ltd; p.342–406.
- International Union for Conservation of Nature (IUCN). 2018a. The IUCN red list of threatened species. Haliaeetus albicilla. Brussels, Belgium: IUCN; Version 3.1, [accessed 2018 Oct 14]. http://www.iucnredlist.org.
- International Union for Conservation of Nature (IUCN). 2018b. The IUCN red list of threatened species. Pernis ptilorhynchus. Brussels, Belgium: IUCN. Version 3.1, [accessed 2018 Oct 14]. http://www.iucnredlist.org.
- Jeon HS, Myeong H, Kang SG, Kim JA, Lee SH, Lee MY, An J. 2018. The mitochondrial genome of Milvus migrans (Aves, Accipitriformes, Accipitridae), an endangered species from South Korea. Mitochondrial DNA B. 3:498–499.
- Laslett D, Canback B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Lerner HRL, Mindell DP. 2005. Phylogeny of eagles, old world vultures, and other accipitridae based on nuclear and mitochondrial DNA. Mol Phylogenet Evol. 37:327–346.
- Mindell DP, Sorenson MD, Dimcheff DE. 1998a. Multiple independent origins of mitochondrial gene order in birds. Proc Natl Acad Sci USA. 95:10693–10697.
- Mindell DP, Sorenson MD, Dimcheff DE. 1998b. An extra nucleotide is not translated in mitochondrial ND3 of some birds and turtles. Mol Biol Evol. 15:1568–1571.
- President of National Institute of Biological Resources (PNIBR). 2017. The guidebook of Korean endangered wildlife. Korea (In Korean)
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
