Abstract
Dysphania pumilio (R.Br.) Mosyakin & Clemants which belongs to Amaranthaceae is an invasive species all over the world. In this study, we presented first complete chloroplast genome of D. pumilio of which length is 151,962 bp consisting four subregions: 83,758 bp of large single copy (LSC) and 17,742 bp of small single copy (SSC) regions are separated by 25,231 bp of inverted repeat (IR) regions. One hundred and twenty-eight genes (84 protein-coding genes, eight rRNAs, and 36 tRNAs) were annotated successfully. The overall GC content of the chloroplast genome is 36.9% and those in the LSC, SSC and IR regions are 34.8%, 30.4%, and 42.7%, respectively. First chloroplast genome of Dysphania will provide accurate phylogenetic position of Dysphania genus among neighbour genera.
Dysphania R. Br. containing aromatic plant species has been traditionally applied to 7–10 species which were endemic to Australia (Aellen Citation1930; Scott Citation1978; Wilson et al. Citation1983). From APG III system, genus Dysphania belonged into Amaranthaceae because Chenopodiaceae was merged into Amaranthaceae (Angiosperm Phylogeny Group Citation2009). Dysphania is clearly distinguished from Chenopodium by multicellular glandular hairs (Judd and Ferguson Citation1999). Recent taxonomic treatments have expanded this genus by integrating Suckleya, Cycloloma, and Ambrosia resulting up to 45 species (Herbarium WA, Citation1998–2009; Mosyakin and Clemants Citation2002; Verloove and Lambinon Citation2006; Mosyakin and Clemants Citation2008). Dysphana pumilio (R.Br.) Mosyakin & Clemants is one of the popular invasive species, which has already been spread out in many countries including Korea (Kang and Shim Citation2002), Japan (Mito and Uesugi Citation2004), China (Zhu et al. Citation2007), Czech Republic (Moravcova et al. Citation2010), and Central Europe (Pysek et al. Citation1998) since 1979. In Korea, it was first reported as an unrecorded species in the parking lot of Songnisan National Park in 1992 (Chung et al. Citation2001) and has been spread to many cities through vehicles.
To understand genetic background of an invasive species, D. pumilio, we collected D. pumilio in Gangseo-gu, Seoul, Korea (Voucher in InfoBoss Cyber Herbarium (IN); IB-00568). Total DNA was extracted from fresh leaves of D. pumilio by using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using HiSeq4000 at Macrogen Inc., Korea, and de novo assembly was done by Velvet 1.2.10 (Zerbino and Birney Citation2008). All bases were confirmed by alignment results generated by BWA 0.7.17 (Li Citation2013) and SAM tools 1.9 (Li et al. Citation2009). Geneious R11 11.0.5 (Biomatters Ltd., Auckland, New Zealand) was used for chloroplast genome annotation.
The chloroplast genome of D. pumilio (GenBank accession is MH936550) is 151,962 bp in length and has four subregions: 83,758 bp of large single copy (LSC) and 17,742 bp of small single copy (SSC) regions are separated by 25,231 bp of inverted repeat (IR). It contains 128 genes (84 protein-coding genes, eight rRNAs, and 36 tRNAs). The overall GC content of D. pumilio is 36.9% and those in the LSC, SSC, and IR regions are 34.8%, 30.4%, and 42.7%, respectively.
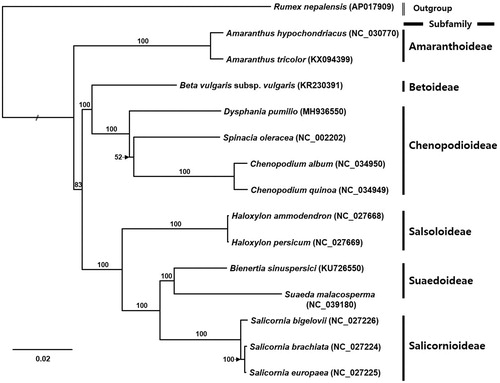
Fourteen complete chloroplast genomes in Amaranthaceae and one species as an outgroup were analysed for constructing phylogenetic tree using maximum likelihood method. Alignment was conducted by MAFFT 7.388 (Katoh and Standley Citation2013) and phylogenetic tree was generated by IQ-TREE 1.6.6 (Nguyen et al. Citation2014). The tree presents that Dysphania genus is distinctly delimitated from Chenopodium which were considered as one genus (). Our first Dysphania chloroplast genome will be utilized to understand accurate phylogenetic relationship among three genera, Dysphania, Spinacia, and Chenopodium, with upcoming additional chloroplast genomes of these genera.
Figure 1. Maximum likelihood phylogenetic tree of Amaranthaceae based on 15 complete chloroplast genomes: Dysphania pumilio (MH936550 in this study), Rumex nepalensis (AP017909), Amaranthus hypochondriacus (NC_030770), Amaranthus tricolor (KX094399), Beta vulgaris subsp. vulgaris (KR230391), Spinacia oleracea (NC_002202), Chenopodium album (NC_034950), Chenopodium quinoa (NC_034949), Haloxylon ammodendron (NC_027668), Haloxylon persicum (NC_027669), Bienertia sinuspersici (KU726550), Suaeda malacosperma (NC_039180), Salicornia bigelovii (NC_027226), Salicornia brachiata (NC_027224), and Salicornia europaea (NC_027225). The numbers above branches indicate bootstrap support values.
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- Aellen P. 1930. Die Wolladventiven Chenopodien Europas. Verh Naturf Ges Basel. 41:77–104.
- Angiosperm_Phylogeny_Group. 2009. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc. 161:105–121.
- Chung Y, Lee C, Paik W-K, Ahn J-C, Park A-E. 2001. Taxonomical investigation of the goosefoot (Genus Chenopodium; Chenopodiaceae) in Korea on a basis of external morphological characters. Kor J Weed Sci. 21:229–235.
- Judd WS, Ferguson I. 1999. The genera of Chenopodiaceae in the southeastern United States. Harvard Pap Bot. 4:365–416.
- Kang B-H, Shim SI. 2002. Overall status of naturalized plants in Korea. Kor J Weed Sci. 22:207–226.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Preprint arXiv. 13033997.
- Herbarium WA. 1998–2009: Flora-Base—The Western Australian Flora. Department of Environment and Conservation.
- Mito T, Uesugi T. 2004. Invasive alien species in Japan: the status quo and the new regulation for prevention of their adverse effects. Glob Environ Res. 8:171–193.
- Moravcova L, Pyšek P, Jarošík V, Havlíčková V, Zákravský P. 2010. Reproductive characteristics of neophytes in the Czech Republic: traits of invasive and non-invasive species. Preslia. 82:365–390.
- Mosyakin S, Clemants S. 2002. New nomenclatural combinations in Dysphania R. Br.(Chenopodiaceae): taxa occurring in North America. Ukr Bot Zhur. 59:380–385.
- Mosyakin SL, Clemants SE. 2008. Further transfers of glandular-pubescent species from Chenopodium subg. Ambrosia to Dysphania (Chenopodiaceae. J Bot Res Inst Texas. 425:431.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2014. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Pysek P, Prach K, Mandák B. 1998. Invasions of alien plants into habitats of Central European landscape: an historical pattern. Plant invasions: ecological mechanisms and human responses; p.23–32.Backhuys.
- Scott A. 1978. A review of the classification of Chenopodium L. and related genera (Chenopodiaceae). Bot Jahrb. 100:205–220.
- Verloove F, Lambinon J. 2006. The non-native vascular flora of Belgium: a new nothospecies and three new combinations. Syst Geogr Plants. 76(2):217–220.
- Wilson HD, Barber SC, Walters T. 1983. Loss of duplicate gene expression in tetraploid Chenopodium. Biochem Syst Ecol. 11:7–13.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhu C-s, Tian C-y, Lu S-f, Zhang Y-x, Xu X, He Y-x, Shi S-f. 2007. Investigation on and statistical analysis of alien invasive plants in Henan Province. J Henan Agric Univ. 2:013.
