Abstract
Argopistes tsekooni is a leaf-mining pest on Oleaceae plants in China. The complete mitochondrial genome of A. tsekooni was sequenced and annotated. It had a total length of 16,552 bp, including 13 PCGs, 2 ribosomal RNAs, 22 tRNAs, and a 2020 bp long non-coding AT-rich region. With a total length of 11,140 bp, all 13 PCGs started with ATN, 9 PCGs terminated with TAA, 3 PCGs terminated with TAG, except that COX3 terminated with an incomplete stop codon T. Maximum-likelihood phylogenetic analysis showed that A. tsekooni was closely grouped with seven other Galerucinae species, which did not contradict the previous morphological and molecular classification.
Argopistes tsekooni is an important garden pest that widely distributes in China. Their larvae are internal-feeding leaf-miners but their adults feed outside and make leaf holes. When outbreak, the beetle could seriously damage Oleaceae species, such as privets (Ligustrum spp), lilacs (Syringa spp), and sweet olive (Osmanthus fragrans) (Liu et al. Citation2018b). A. tsekooni individuals were collected in 6 March 2018 at Gannan Normal University, Jiangxi Province, China (N 25.798, E 114.888). All specimens were deposited at Leafminer Group, School of Life Sciences, Gannan Normal University (GNNU). Adults were stored at −80 °C in 100% ethanol. In this study, the complete mitochondrial genome of A. tsekooni (GenBank accession number MK060107) was sequenced and annotated according to the Illumina pair-end sequencing data. Although Liu et al. (Citation2018b) explored the phylogenetic relationship of A. tsekooni with other leaf beetles based on COX1 (COI), it is the first report on Argopistes phylogeny based on complete mitogenomes.
The complete circular mitogenome of A. tsekooni was 16,552 bp (A + T = 79.52%, A = 41.51%, T = 38.01%, G = 8.32%, C = 12.16%), and included 13 PCGs, 2 ribosomal RNAs, 22 tRNAs, and a 2020 bp long non-coding AT-rich region (A + T = 82.2%). The arrangement of the mitochondrial genes was not different with the most common type of insect putative ancestors (Boore Citation1999; Cameron Citation2014). For the 37 genes, 4 PCGs (NAD 1, 4, 4L, 5), 2 rRNAs (rRNA L, S), and 8 tRNAs (tRNA Q, C, Y, F, H, P, L1, V) were located in J-strand (minus strand), while the other 9 PCGs and 14 tRNAs in N-strand (plus strand). With a total length of 11,140 bp, all 13 PCGs started with ATN (ATA for COX2 and NAD6; ATG for ATP6, COX3, NAD4, NAD4L, and COB; ATT for NAD2, COX1, ATP8, NAD3, NAD5, and NAD1). And 9 PCGs terminated with TAA, 3 PCGs (NAD 1, 3, COB) terminated with TAG, except that COX3 terminated with an incomplete stop codon T. The longest one was NAD5 gene (1716 bp) and the shortest one was ATP8 gene (156 bp). The rRNAL was 1290 bp long with an A + T content of 82.6% and the rRNAS was 739 bp long with an A + T content of 73.4%. All 22 tRNAs were ranging from 63 to 71 bp and had a typical clover-leaf secondary structure, but tRNAS1 (AGN) lacked a stable dihydrouridine (DHU) stem (Guo et al. Citation2017; Xu et al. Citation2018).
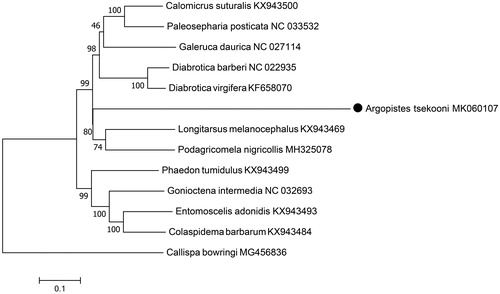
We downloaded mitogenome sequences of 12 Chrysomelidae species from GenBank which including seven Galerucinae species, four Chrysomelinae species, and one Cassidinae species. The phylogenetic tree was constructed in MEGA version 7.0 (Kumar et al. Citation2016) using maximum likelihood method (Stamatakis Citation2014), with Callispa bowringi (Jiang et al. Citation2018; Liu et al. Citation2018a) as the outgroup (). A. tsekooni was closely grouped with seven other Galerucinae species, which did not contradict the previous morphological and molecular classification (Liu et al. Citation2018b).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59:95–117.
- Guo Q, Xu J, Liao C, Dai X, Jiang X. 2017. Complete mitochondrial genome of a leaf-mining beetle, Agonita chinensis Weise (Coleoptera: Chrysomelidae). Mitochondrial DNA Part B Resour. 2:532–533.
- Jiang X, Guo Q, Xu J, Liu P, Long C, Dai X. 2018. Complete mitochondrial genome of a leaf-mining beetle, Podagricomela nigricollis (Coleoptera: Chrysomelidae). Mitochondrial DNA Part B Resour. 3:721–722.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Liu P, Guo Q, Xu J, Liao C, Dai X. 2018a. Complete mitochondrial genome of a leaf beetle, Callispa bowringi (Coleoptera: Chrysomelidae). Mitochondrial DNA Part B Resour. 3:213–214.
- Liu P, Zhong M, Xu J, Guo Q, Liao C, Dai X. 2018b. Life history and phylogenetic analysis of Argopistes tsekooni. North Horticult. 42:59–63.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Xu J, Liao C, Guo Q, Long C, Dai X. 2018. Mitochondrial genome of a leaf-mining beetle Prionispa champaka Maulik (Coleoptera: Chrysomelidae: Cassidinae). Mitochondrial DNA Part B Resour. 3:147–148.