Abstract
Rhizophora stylosa is a true mangrove distributed in the Indo-Pacific region. In this study, the whole chloroplast genome of R. stylosa was assembled and annotated. The chloroplast genome was 164,475 bp in length, consisting of a large single copy (LSC) region of 92,582 bp, a small single copy (SSC) region of 19,243 bp, and two inverted repeat (IR) regions of 26,325 bp. It contained 131 genes, including 85 protein-coding genes, 38 tRNA genes, and eight rRNA genes. The overall GC content was 34.9%. Phylogenetic analysis showed that R. stylosa was sister to Erythroxylum novogranatense.
Mangroves species have significant ecosystem functions, such as coastal protection, land stabilization, and water purification. However, they are at high risk of disappearance due to human activities and climate change (Romañach et al. Citation2018). Rhizophora stylosa Griff. (Rhizophoraceae) is distributed in the Indo-Pacific region, extending in the Western Pacific from South West Rocks to Fiji and the Marshall Islands and north to the Ryukyu Islands, and likely with a western limit of the Andaman and Nicobar Islands (Vannucci Citation2002; Wilson and Saintilan Citation2012). Rhizophora stylosa is commonly found in open seawater or at the seaward edge of estuaries, thus it is one of the most vulnerable mangrove species to climate change and rising sea-level. However, due to habitat destruction and removal of mangrove areas, there has been an approximately 20% decline within the species range since 1980 (FAO Citation2007). Rhizophora stylosa is therefore listed in the IUCN Red List of Threatened Species (Ellison et al. Citation2010). In this study, we characterized the complete chloroplast (cp) genome of R. stylosa to better understand scientific conservation strategies for this species.
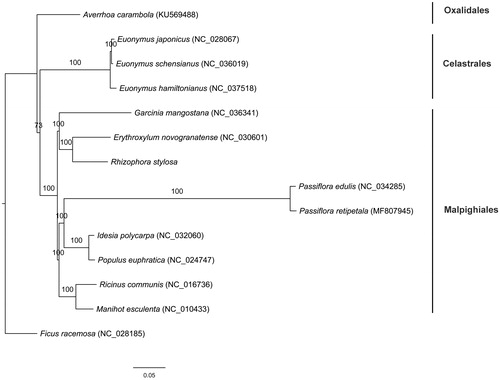
The plant material of R. stylosa was sampled from Hepu, Guangxi, China (N21°29′53″, E109°45′39″). The voucher specimen (ZhouLX13701) was deposited at South China Botanical Garden Herbarium. Total genome DNA was extracted from fresh leaves using a modified CTAB protocol (Doyle and Doyle 1987). Library construction and paired-end sequencing were performed by Beijing Genomics Institute (Shenzhen, China). Genome sequences were assembled in NOVOPlasty (Dierckxsens et al. Citation2017). The genes in the chloroplast genome were annotated using GeSeq (Tillich et al. Citation2017), with manual adjustment using Geneious ver. 11.0.2 (Kearse et al. Citation2012). Phylogenomic analysis was performed using the complete cp genomes from 13 species in the orders Malpighiales (including R. stylosa in this study), Celastrales, and Oxalidales. Ficus racemosa from Rosales was used as outgroup species. The cp genome sequences were aligned using MAFFT (Katoh and Standley Citation2013). The phylogenetic tree was constructed with RAxML (Stamatakis Citation2014) implemented in Geneious, using the maximum likelihood algorithm.
The R. stylosa cp genome (GenBank accession no.: MK070169) is 164,475 bp in length, consisting of a large single copy (LSC) region of 92,582 bp, a small single copy (SSC) region of 19,243 bp, and two inverted repeat (IR) regions of 26,325 bp. The overall GC content was 34.9%, higher than those of LSC (32.0%) and SSC (28.6%) regions, but lower than those of IR regions (42.2%). The chloroplast genome of R. stylosa contained 131 genes, including 85 protein-coding genes, 38 tRNA genes, and eight rRNA genes. Among the protein-coding genes, ten genes contain a single intron while four genes possess two introns. Most genes occurred in single copy, while eight protein-coding genes, eight tRNA genes, and four rRNA genes in IR regions were duplicated. Based on the ML tree, all sampled members of Malpighiales formed a clade, and R. stylosa was sister to Erythroxylum novogranatense (). The chloroplast genome of R. stylosa is of significance for its conservation and evolutionary studies.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- FAO. 2007. The World's Mangroves 1980-2005. FAO Forestry Paper 153. Rome: Forestry Department, Food and Agriculture Organization of the United Nations (FAO).
- Ellison J, Duke N, Kathiresan K, Salmo III SG, Fernando ES, Peras JR, Sukardjo S, Miyagi T. 2010. Rhizophora stylosa. The IUCN Red List of Threatened Species 2010. e.T178850A7626520.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Romañach SS, DeAngelis DL, Koh HL, Li Y, Teh SY, Barizan RSR, Zhai L. 2018. Conservation and restoration of mangroves: global status, perspectives, and prognosis. Ocean Coast Manag. 154:72–82.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes . Nucleic Acids Res. 45:W6–W11.
- Vannucci M. 2002. Indo-west pacific mangrove. In: Lacerda LD, editor. Mangrove ecosystems: function and management. Berlin: Springer; p. 123–215.
- Wilson NC, Saintilan N. 2012. Growth of the mangrove species Rhizophora stylosa Griff. at its southern latitudinal limit in eastern Australia. Aquat Bot. 101:8–17.