Abstract
Rubus is one large and diverse genus in the rose family, Rosaceae, comprising of about 750 species distributed across the world. Hybridization, polyploidization, and agamospermy complicate the taxonomy within the genus, and further challenge the utilization and improvement of these plants as crops. Herein, we report the complete chloroplast genome of R. leucanthus, assembled from genome resequencing data using Illumina paired-end reads. The assembled chloroplast genome was 156,068 bp in length, consisting of a large single-copy (LSC) region of 85,749 bp, a small single-copy (SSC) region of 18,763 bp, separated by two inverted repeat (IR) regions of 25,778 bp each. A total of 131 genes were predicted, with an overall GC content of 37.03%. Phylogenetic analysis placed R. leucanthus in the tribe Rubeae, well separated from other tribes in the Rosaceae family.
Rubus is one large and diverse genus in the family Rosaceae, comprising of about 750 species distributed around the world (Robertson Citation1974; Thompson Citation1995). By virtue of its medicinal and cosmetic values, some of the wild Rubus species have been domesticated. Besides highly variable morphologies, extensive hybridization, polyploidization, and agamospermy also complicate the classification within this genus (Alice and Campbell Citation1999). A well-resolved phylogeny based on sufficient molecular markers is essential to understand the relationships between species, and the efficient utilization and improvement of these wild Rubus species as crop. To this end, we report the complete chloroplast genome sequence of the white-flowered raspberry R. leucanthus Hance, a wild species widespread in subtropical and tropical China, as a resource for future studies on the taxonomy of Rubus.
An individual plant of R. leucanthus was sampled from Huadu, Guangzhou, China, and the voucher specimen (RLEU-2018-GZ01) is kept at the Sun Yat-sen University Herbarium (SYS). Total genomic DNA was obtained and subjected to library construction and sequencing on an Illumina Hiseq2500 platform. Using approximately 4 Gb of paired-end (125 bp) reads and a conserved rbcL gene sequence (GenBank accession AWQ64348.1) as input, we assembled the chloroplast genome of R. leucanthus via a seed-and-extend algorithm implemented in NOVOPlasty (Dierckxsens et al. Citation2017). In addition, eight randomly selected genes were Sanger-sequenced to verify the accuracy of the automated assembly. DNA sequences of the seven genes (rps16, atpF, rpoC2, rpoC1, atpE, matK, and ndhD), covering approximately 10,458 bp (6.7% of the total chloroplast genome size), had 100% match to those on the assembled genome. Using Verdant (McKain et al. Citation2017) and DOGMA (Wyman et al. Citation2004), the chloroplast genome was annotated with manual corrections.
The length of the complete chloroplast genome sequence of R. leucanthus (GenBank accession MK105853) was 156,068 bp, with a large single-copy (LSC) region of 85,749 bp, a small single-copy (SSC) region of 17,094 bp, separated by two inverted repeat (IR) regions of 25,778 bp each. A total of 131 genes were predicted, including 86 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The overall GC content was 37.03%. The petD gene had an AUC start codon, instead of the conventional AUG start codon.
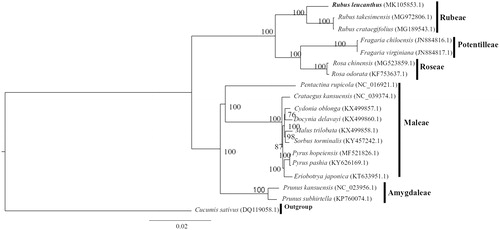
For phylogenetic tree construction, the chloroplast genomes of R. leucanthus, 17 other Roseaceae species, and one outgroup species Cucumis sativus (DQ119058.1) downloaded from GenBank were aligned using MAFFT v7.307 (Katoh & Standley Citation2013). A maximum likelihood tree () was then constructed using RAxML (Stamatakis, Citation2014). Rubus leucanthus was placed in the Rubeae tribe, showing high support for monophyly among the five tribes used in this study.
Acknowledgements
We thank Yubing Zhou for assistance in using NOVOPlasty, and Verdant.
Disclosure statement
The authors declare that there is no conflict of interest regarding the publication of this article. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Alice L, Campbell C. 1999. Phylogeny of Rubus (rosaceae) based on nuclear ribosomal DNA internal transcribed spacer region sequences. Am J Bot. 86:81–97.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- McKain MR, Hartsock RH, Wohl MM, Kellogg EA. 2017. Verdant: automated annotation, alignment and phylogenetic analysis of whole chloroplast genomes. Bioinformatics. 33:130–132.
- Robertson K. 1974. The genera of Rosaceae in the southeastern United States. J Arnold Arboretum. 55:352–360.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Thompson M. 1995. Chromosome numbers of Rubus species at the National Clonal Germplasm Repository. HortScience. 30:1447–1452.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.