Abstract
The study has determined the complete mitochondrial genome sequence of the Rhabdophis adleri. The mitogenome was 17,505 bp in size, containing 26.33% T, 27.08% C, 33.38% A, and 13.21% G, 2 ribosomal RNA (rRNA) genes, 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, as well as 2 Dloop regions. The gene arrangement, nucleotide composition, and the codon usage pattern of the mitochondrial genome of R. adleri were alike in other alethinophidian snakes. The phylogenetic reconstruction analyses yielded similar topologies, having been expressed that R. adleri can strongly be placed within Colubridae.
Rhabdophis adleri is an endemic snake species of Hainan, China (Zhao Citation1997), has since been considered a terrestrial wild animal with important economic and scientific value since 2000 (Li Citation2011). The validity of R. adleri was explored by morphological characteristics, distribution (Zhao Citation1997), hemipenial morphology (Zhu et al. Citation2013), nuchal and nucho-dorsal glands (Mori et al. Citation2016), but little is known to the complete mitochondrial DNA of R. adleri. Mitochondrial DNA is widely used molecular marker for estimating phylogenetic relationships in animals (Lin and Danforth Citation2004; Gissi et al. Citation2008). In present study, the complete mitogenome of R. adleri was sequenced and annotated, its main features were provided. We further discussed the phylogenetic relationships of the snakes based on the protein-coding sequences.
The specimen of R. adleri (ZGX125, Zhu GX Field tag, stored in the zoological specimens room, College of Life Science, SICAU) was collected from Five Finger Mountain (Mt. Wuzhi, 18°43′41″N, 109°51′48″E), Hainan Province. The total genomic DNA was extracted from the muscle of R. adleri using Genomic DNA Extraction Kit according to the instruction. Sixteen pairs of primers were performed to amplify contiguous, overlapping segments covering the complete mitogenome of R. adleri. The mitogenome sequence and protein-coding genes were assembled and analyzed by MEGA 5.0 (Tamura et al. Citation2011). Phylogenetic analysis by BI was carried out using MrBayes 3.1.2 (Huelsenbeck and Ronquist Citation2001) and ML analysis was performed using PAUP4.0b10 (Swofford Citation2003) ().
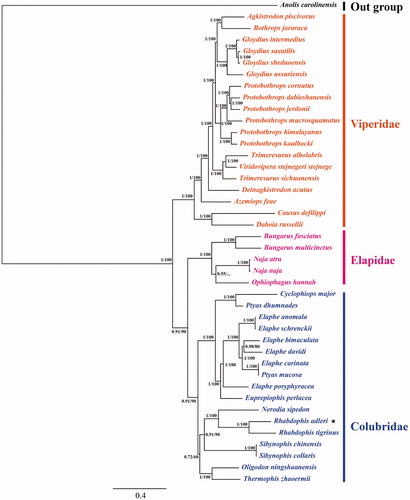
Figure 1. Molecular phylogenetic tree of 42 snake species. Values on the nodes indicate bootstrap and posterior probability support (BS/PP), ★ indicated Rhabdophis adleri.
Total length of the R. adleri mitochondrial genome was 17,505 bp, and it was a typically circular molecular. Total base composition was 26.33% T, 27.08% C, 33.38% A, and 13.21% G. It contained 37 typical mitochondrial genes (13 PCGs, 22 tRNAs, and 2 rRNAs) and two control regions (Dloop1 and Dloop2). Most mitochondrial genes were encoded on the H-strand, except for the ND6 and nine tRNA genes: tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer (UCN), tRNAGlu, tRNAThr, and tRNAPro, which were encoded on the L-strand. The gene order, orientation and nucleotide composition were similar to the previously published Colubrinae mitogenomes (Li et al. Citation2016; Peng et al. Citation2016; Weng et al. Citation2016). The 12S rRNA(928 bp) was located between tRNAPhe and tRNAVal, while 16 S rRNA (1447 bp) was between tRNAVal and ND1. Dloop1 was located between tRNAPro and tRNAPhe, while Dloop2 was between tRNAIle and tRNALeu (UUR). Like most snakes in the current study, the mitochondrial genome of R. adleri contained a duplicated Dloop region between ND1 and ND2, except some scolecophidian snakes that had only one Dloop region (He et al. Citation2010).
The phylogenetic position of R. adleri was constructed using the 12 concatenated protein-coding genes. The results indicated that all analyses produced four major groups, the Iguanidae (the out group), Colubridae, Elapidae and Viperidae (). Rhabdophis adleri and R. tigrinus formed a single monophyletic group, and belonged to Colubridae (Zhao Citation1997; Zhao et al. Citation2016). Our study first revealed complete mitochondrial genome of R. adleri, which can contribute to the conservation of this important economic and scientific wild animal’s genetic resources.
Acknowledgements
We thank Dr. Jichao Wang and Ling Ding for their assistance in sample collection.
Disclosure statement
The authors declare that they have no competing interests. The authors alone are responsible for writing of this paper.
Additional information
Funding
References
- Gissi C, Iannelli F, Pesole G. 2008. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity. 101:301.
- He M, Feng J, Zhao E. 2010. The complete mitochondrial genome of the Sichuan hot-spring keel-back (Thermophis zhaoermii; Serpentes: Colubridae) and a mitogenomic phylogeny of the snakes. Mitochondrial DNA. 21:8–18.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
- Li HM. 2011. The distribution and perniciousness of Rhabdophis subminiatus at Ailaoshan in Xinping County. Hubei Agri Sci. 50:800–801.
- Li E, Sun F, Zhang R, Chen J, Wu X. 2016. The complete mitochondrial genome of the striped-tailed rat-snake, Orthriophis taeniurus (Reptilia, Serpentes, Colubridae). Mitochondrial DNA. 27:599–600.
- Lin CP, Danforth BN. 2004. How do insect nuclear and mitochondrial gene substitution patterns differ? Insights from Bayesian analyses of combined datasets. Mol Phylogenet Evol. 30:686–702.
- Mori A, Jono T, Ding L, Zhu G, Wang J, Shi H, Tang Y. 2016. Discovery of nucho-dorsal glands in Rhabdophis adleri. Current Herpetology. 35:53–58.
- Peng LF, Weng SY, Yang DC, Lu CH, Huang S. 2016. Complete mitochondrial genome of the Xianggelila Hot-spring snake, Thermophis shangrila (Reptilia, Colubridae). Mitochondrial DNA Part B Resources. 1:536–537.
- Swofford DL. 2003. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4.0b10. Sunderland, MA: Sinauer Associates.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Weng SY, Peng LF, He M, Duan SQ, Huang S. 2016. The complete mitochondrial genome of the Xizang hot-spring snake, Thermophis baileyi Wall, 1907 (Reptilia, colubridae). Mitochondrial DNA Part B. 1:921–922.
- Zhao D, Liu H, Zhao WG, Liu P. 2016. The complete mitochondrial DNA sequence and the phylogenetic position of Rhabdophis tigrinus (Reptilia: Squamata). Mitochondrial DNA Part B. 1:216–217.
- Zhao EM. 1997. A new species of Rhabdophis (Serpentes: Colubridae) from Hainan Island, China. Asiatic Herpetol Res. 7:166–169.
- Zhu GX, Guo P, Zhao EM. 2013. Comparative studies on hemipenial morphology of eight species of Keelback snakes (Serpentes: Colubridae: Rhabdophis). Sichuan J Zool. 32:380–384.
