Abstract
Dipelta yunnanensis (Caprifoliaceae) is a medicinal plant endemic to China. It has been included in the Chinese Species Red List and is also listed as a vulnerable species in the IUCN Red List. In this study, we determined the complete chloroplast (cp) genome sequence of Dipelta yunnanensis using Illumina paired-end sequencing. The whole cp genome was 155,359 bp in length, consisting of a pair of inverted repeat regions of 23,397 bp, a small single-copy region of 19,098 bp, and a large single-copy region of 89,467 bp. The genome encoded 128 unique genes including 82 protein-coding genes, 37 tRNA genes, and eight rRNA genes. The overall GC content of the genome was 38.4%. Phylogenetic analysis with the reported chloroplast genomes of Dipsacales revealed that D. yunnanensis had a close relationship with Kolkwitzia amabilis. The chloroplast genome sequence of D. yunnanensis offers a useful resource for future conservation genetics and phylogenetic studies.
Dipelta yunnanensis is a deciduous shrub belonging to the genus Dipelta of family Caprifoliaceae (Yang and Landrein Citation2011) and mainly distributed in Yunnan, Hubei, Sichuan, China. It is of great value in landscape design and its root is used as a traditional Chinese medicine to treat measles, pruritus, and Damp-Heat syndrome (Wang Citation2014). Recently, the natural populations of D. yunnanensis have been declining dramatically due to the excessive deforestation and continuous deterioration of habitats. It has been classified as a vulnerable plant listed in the category of the Red List of biodiversity in China and International Union for Conservation of Nature and Natural Resources (http://rep.iplant.cn/prot/Dipelta%20yunnanensis). To date, only SSR and AFLP markers have been developed to identify the interspecific relationships in Dipelta (Liu et al. Citation2013), and genomic information of the genus is lacking for further study. Here we characterized the complete chloroplast genome of D. yunnanensis based on the whole-genome Illumina sequencing dataset to obtain genomic information for conservation studies and to explore its phylogenetic placement within Dipsacales.
Fresh leaves of D. yunnanensis were collected from Shangri-la, (Yunnan, China; 100.05°E, 27.19°N), and the voucher (2018JH15) was deposited at the Evolutionary Botany Laboratory (EBL), Northwest University. Genomic DNA isolated with DNAquick Plant System (Tiangen Biotech (Beijing) CO. LTD) was sequenced using the Illumina HiSeq 2500 platform in Biomarker Technologies CO, China. The raw reads were trimmed using NGSQC Toolkit_v.2.3.3 with the default parameters (Patel and Jain Citation2012). Subsequently, the clean reads were used to perform referenced-guided assembly using MIRA 4.0.2 (Chevreux et al. Citation2004) and MITObim v1.8 (Hahn et al. Citation2013) with cp genome of Dipelta floribunda (NC_037955) as the reference. Assembled plastome was annotated using Geneious R v9.0.5 (Kearse et al. Citation2012) by aligning to the reference plastome. The consensus sequences were imported into the online program Dual Organellar Genome Annotator DOGMA (Wyman et al. Citation2004) for gene annotation.
The complete chloroplast genome of D. yunnanensis (GenBank accession no. MK002702) was 155,359 bp in length, comprising of a large single copy (LSC) region of 89,467 bp, a small single copy (SSC) region of 19,098 bp, and a pair of inverted repeat (IR) regions of 23,397 bp. The overall GC content was 38.4% with the corresponding values in the LSC, SSC, and IR region of 36.5%, 33.1%, and 44.2%, respectively. The chloroplast genome contained 128 genes including 82 protein-coding genes, 37 tRNA genes, and 8 rRNA genes.
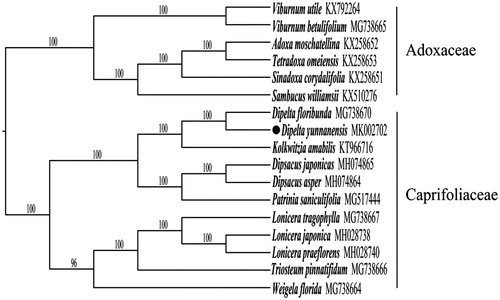
In order to understand the phylogenetic relationship between D. yunnanensis and its related species, we selected 10 published cp genomes of Caprifoliaceae and six Adoxaceae species as outgroup. The whole plastome sequences were aligned with MAFFT 7.310 (Katoh and Standley Citation2016), and the maximum likelihood (ML) tree was reconstructed by RAxML v 7.2.8 (Stamatakis Citation2006) with 1000 bootstrap replicates. The result indicated that D. yunnanensis was sister to D. floribunda and closely related to Kolkwitzia amabilis. Both of the two genera are classified into the new genus Linnaea (Christenhusz Citation2013) ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Müller WE, Wetter T, Suhai S. 2004. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14:1147–1159.
- Christenhusz MJM. 2013. Twins are not alone: a recircumscription of Linnaea (Caprifoliaceae). Phytotaxa. 125:25–32.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads: a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Katoh K, Standley DM. 2016. A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics. 32:1933–1942.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxt S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Liu ZL, Tian P, Li JF, Guo C, Biffin E. 2013. A comparison of DNA sequences, SSR and AFLP for systematic study of Dipelta (caprifoliaceae). Ann Bot Fenn. 50:351–359.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE. 7:e30619
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Wang GQ. 2014. National compilation of Chinese herbal medicine. Beijing (BJ): People's Health Press.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yang Q, Landrein S. 2011. Flora of China. Vol. 19, Linnaeaceae. Beijing: Science Press, and St. Louis: Missouri Botanical Garden Press.