Abstract
Draba oreades is one of the indicator species that narrowly distributed in alpine habitats. Here, the complete plastid genome (plastome) of D. oreades was determined by Illumina paired-end sequencing technology. It was 153,503 bp in length with a large single copy region of 82,683 bp, a small single copy region of 18,094 bp, and a pair of inverted repeat regions of 26,363 bp. A total of 129 genes were identified, including 84 protein-coding genes (78 PCG species), 37 tRNA genes (30 tRNA species), and 8 rRNA genes (4 rRNA species). The overall GC content of D. oreades plastome is 36.5%. Besides, we totally detected 116 perfect simple sequence repeats in D. oreades plastome. Maximum likelihood phylogenetic analysis based on 27 complete plastome sequences revealed that D. oreades was closely related to its congeneric D. nemorosa.
Draba, the largest genus in Brassicaceae with about 350 species, is distributed in the Arctic, subarctic, alpine regions of the world. The interspecific phylogenetic relationships in the genus still remain unclear due to frequent hybridization and polyploidization (Zhou et al. Citation2001, Jordon-Thaden et al. Citation2010). In this study, the complete plastid genome of Draba oreades, a perennial herb in alpine mountains of western China, was generated with the Illumina paired-end sequencing technology to provide valuable information for phylogenetic and evolutionary studies in both Draba and Brassicaceae taxa.
The samples of D. oreades were collected from Taibai Mountain (Shaanxi, China; 107.77°E, 33.95°N) and the voucher (2017JH23) was deposited at the Evolutionary Botany Laboratory (EBL), Northwest University. The whole-genome sequencing was performed by the Illumina Hiseq 2500 platform in Biomarker Technologies Inc. (Beijing, China). After trimming and filtering low quality reads with NGSQCToolkit v2.3.3 (Patel and Jain Citation2012), 6.39G clean reads were used for the plastome assembly by program MIRA v4.0.2 (Chevreux et al. Citation2004), with that of the congeneric D. nemorosa (NC_009272) as the reference. Then, we used the program MITObim v1.8 (Hahn et al. Citation2013) to further assemble the plastome. Genes were annotated by comparing to the reference plastome and manually adjusted in GENEIOUS R8.0.2 (Biomatters Ltd., Auckland, New Zealand). Genome map was drawn using OrganellarGenome DRAW (OGDRAW) (Lohse et al. Citation2013). The simple sequence repeats (SSRs) were detected using MISA (http://pgrc.ipk-gatersleben.de/misa/) with the minimum repeat unit of 10 for mono-, 5 for di-, and 4 for tri-, and 3 for tetra-, penta-, and hexa-nucleotides, respectively.
The D. oreades plastome (GenBank accession number: KY947352) was 153,503 bp in length with a large single copy (LSC) region of 82,683 bp, a small single copy (SSC) region of 18,094 bp, and two copies of inverted repeat (IR) region of 26,363 bp. It contained 129 genes, including 84 protein-coding genes, 8 ribosomal RNA genes, and 37 tRNA genes. Among these genes, 14 genes harboured a single intron (trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC, atpF, ndhA, ndhB, rpoC1, petB, petD, rpl2, and rpl16) and 3 genes (ycf3, clpP, and rps12) harboured two introns. The overall GC content of D. oreades plastome was 36.5%, with the corresponding values in the LSC, SSC, and IR region of 34.3%, 29.3%, and 42.4%, respectively. A total of 116 perfect SSRs were found in D. oreades plastome, including 80 mono-, 21 di-, 5 tri-, 7 tetra-, 1 penta-, and 2 hexanucleotide repeats. These AT-rich SSRs were mainly located in intergenic or intron regions.
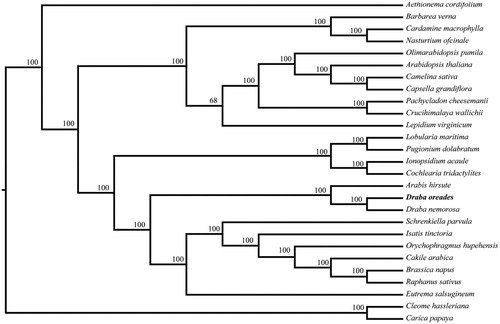
To ascertain its phylogenetic placement within Brassicaceae, maximum likelihood analysis was conducted using RAxMLv7.2.8 (Stamatakis Citation2006) under GTRGAMMA model with 500 replicates. The phylogenetic tree of 25 representatives in Brassicaceae as well as two outgroups, Carica papaya (NC_010323) and Cleome hassleriana (NC_034364) showed that Brassicaceae was a monophyletic clade, as well as D. oreades and its congeneric D. nemorosa had the closest relationship (). The new plastome information of D. oreades will facilitate the population and evolutionary studies of Draba species and the conservation of this alpine herb.
Figure 1. The phylogenetic tree based on 27 complete plastid genome sequences. Accession numbers: Draba oreades KY947352, Aethionema cordifolium NC_009265, Arabidopsis thaliana NC_000932, Arabis hirsute NC_009268, Barbarea verna NC_009269, Brassica napus NC_016734, Cakile arabica NC_030775, Camelina sativa NC_029337, Capsella grandiflora NC_028517, Cardamine macrophylla MF405340, Carica papaya NC_010323, Cochlearia tridactylites NC_029332, Crucihimalaya wallichii NC_009271, Draba nemorosa NC_009272, Eutrema salsugineum NC_028170, Ionopsidium acaule NC_029333, Isatis tinctoria NC_028415, Lepidium virginicum NC_009273, Lobularia maritima NC_009274, Nasturtium officinale NC_009275, Olimarabidopsis pumila NC_009267, Orychophragmus hupehensis NC_033500, Pachycladon cheesemanii NC_021102, Pugionium dolabratum NC_030515, Raphanus sativus NC_024469, Schrenkiella parvula NC_028726, Cleome hassleriana (NC_034364)
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Müller WE, Wetter T, Suhai S. 2004. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14:1147–1159.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129
- Jordon-Thaden I, Hase I, Al-Shehbaz I, Koch MA. 2010. Molecular phylogeny and systematics of the genus Draba (Brassicaceae) and identification of its most closely related genera. Mol Phylogenet Evol. 55:524–540.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7:e30619
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Zhou TY, Lu LL, Yang G, Al-Shehbaz IA, editors. 2001. Brassicaceae. Beijing: Science Press and St. Louis: Missouri Botanical Garden (Wu CY, Raven PH, editors. Flora of China; vol. 8).
