Abstract
The complete mitochondrial genomic sequence of Yellowtail Kingfish Seriola aureovittata was determined in this study. The circular mitogenome contains 16,609 bp, encoding 37 typical animal mitochondrial genes and containing a putative D-loop region. Phylogenetic analysis based on the mitochondrial genomes of S. aureovittata indicated that S. aureovittata and Seriola lalandi are the most closely related species and may be the different geographical populations of one species.
The Yellowtail kingfish, Seriola aureovittata is a large marine pelagic predatory fish species which mainly distributed in the global temperate and subtropical zones. It has large edible value and economic value, which make it being cultured artificially in many countries including China as an important commercial species (Iguchi et al. Citation2012). In recent years, most researches were focused on its mass scale rearing and breeding technology (Joh et al. Citation1995), but few studies conducted on the genome and population genetic structure. This study provides basic data basis for working out the differentiation of different geographical populations and kinship relations of S. aureovittata.
Ten individuals of S. aureovittata were collected from Yellow Sea near Wangjia Island, the southern tip of Liaodong Peninsula, Dalian (39°27'N, 123°6'E) and stored in Yellow Sea Fisheries Research Institute Fish reproduction centre, Qingdao, China (number as YSFRI-Seriola-aureovittata-shi-SF2). The whole genomic DNA was extracted from the muscles tissue of S. aureovittata. The complete mitochondrial genome of S. aureovittata was obtained by the second generation sequencing with PE150 Illumina DNA sequencer (Schuster Citation2008) and spliced with software. The length of the complete mitochondrial genome of S. aureovittata is 16,609 bp (NO. MH211123), comprising the same sat of 37 mitochondrial genes contained 13 protein-coding genes, 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and one putative control region (D-loop). It is same as that in other teleost fishes (Degani Citation2013). There are two kinds of initiation codons. All 13 protein-coding genes start with the typical initiation codon ATG except for the ND5 gene start by ATA codon. Eight protein-coding genes including ND1, ND2, COX1, ATPase 8, ATPase 6, COX3, ND4L, and ND5 were ended with complete termination codons TAA, whereas in protein-coding genes ND3 and ND6 is TAG. The other three genes COX2, ND4, and CytB terminate in an incomplete stop codon T. The uncomplete termination codon T will be completed by posttranscriptional polyadenylation (Ojala et al. Citation1981). Some overlapping reading frames are present between the genes, such as 10 nucleotides were overlapped between ATP8 and ATP6, 1 nucleotide was overlapped between ATP6 and COX3, 7 nucleotides were overlapped between ND4Land ND4, and 4 nucleotides were overlapped between ND5 and ND6. The length of 12S and 16S rRNA genes is 951 and 1699 bp, respectively. And the location of them is between the genes tRNAPhe and tRNALeu(UAA), what separate the two genes (12S rRNA and 16S rRNA) is tRNAVal. The 22 tRNA genes rang in size from 66 to 75 bp. Eight of them that interspersed throughout the genome are in light chain while the other tRNA genes in heavy chain. All the tRNA gene products fold into cloverleaf secondary structures except tRNAGly which fold into cosh secondary structures. The length of putative control region is 921 bp and is located between tRNAPhe and tRNAPro.
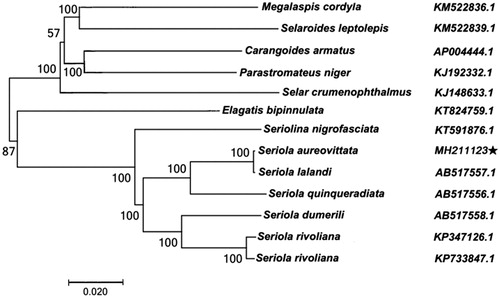
The phylogenetic analysis based on the 13 complete mitochondrial genome data of Carangidae show that S. aureovittata belongs to a Seriola clade and is closely related to Seriola lalandi from Japan ().
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Degani G. 2013. Mitochondrial DNA sequence analysis in Anabantoidei fish. Adv Biol Chem. 03:347–355.
- Iguchi J, Takashima Y, Namikoshi A, Yamashita M. 2012. Species identification method for marine products of Seriola and related species. Fish Sci. 78:197–206.
- Joh YG, Brechany EY, Christie WW. 1995. Characterization of wax esters in the roe oil of amber Fish, Seriola aureovittata. J Am Oil Chem Soc. 72:707–713.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290:470–474.
- Schuster SC. 2008. Next-generation sequencing transforms today's biology. Nat Methods. 5:16–18.