Abstract
In this study, we sequenced and uploaded the complete mitochondrial genome of Daurian pika (Ochotona dauurica) for the first time using tissue obtained from a wild female sample captured from Inner Mongolia, China. This mitochondrial genome is a circular molecule of 17,293 bp in length, consisting of two ribosomal RNA genes, 22 transfer RNA genes, 13 protein-coding genes, 1 control region, and 1 rep_origin region. The phylogenetic analysis basis of 12 protein-coding genes except for ND6 gene shows that Ochotona dauurica clusters with Ochotona curzoniae.
The Ochotona dauurica is classified under order Lagomorpha, family Ochotonidae and genus Ochotona and widely distributed in the typical steppes of central Inner Mongolia (Zhong et al. Citation2008). In this study, the tissue sample of O. dauurica was collected through field survey in Hulun Lake National Nature Reserve, Inner Mongolia, China, and the geo-spatial coordinates are 48°22′16′′N latitude and 117°31′54′′E longitude. The sample died a natural death and was stored at Hulun Lake National Nature Reserve’s herbarium and was frozen in ultra-low temperature freezer. The DNA was extracted with the DNeasy Blood& Tissue kit (QIAGEN, Germantown, MD) . All sampling procedures and experimental manipulations held the proper permits. After manual assembled and annotated, the genome was deposited in GenBank with the accession number MK105852.
The complete mitochondrial genome sequence of O. dauurica is a circular molecule of 17,293 bp in length, consisting of two ribosomal RNA genes, 22 transfer RNA genes, 13 protein-coding genes, 1 control region, and 1 rep origin region. Among these genes, nine genes (ND6, tRNAAsn, tRNASer, tRNAAla, tRNAGlu, tRNACys, tRNATyr, tRNAPro, and tRNAGln) encoded in L-strand and other genes encoded in H-strand. The base composition is 31.0% for A, 29.4% for T, 13.4% for G, 26.2% for C, and the percentage of A and T (60.4%) is higher than G and C (39.6%). The gene arrangement and content are similar to the complete mitochondrial genome of other Ochotona species (Lin et al. Citation2002).
Phylogenetic relationships of O. dauurica and 12 other species were analysed using Bayesian inference (BI) methods maximum-likelihood (ML) based on 12 protein-coding genes except ND6 and the C. lupus (KF857179) was chosen as an out-group. According to the AIC criterion, GTR + I+G was selected as the best-fitting nucleotide substitution model using MrModeltest version 3.7 (Uppsala University, Uppsala, Sweden; Ronquist and Huelsenbeck et al. Citation2003) (Nylander Citation2004). These parameters were used in BI and the ML analysis by MrBayes version 3.2.2 (Uppsala, Sweden; Ronquist and Huelsenbeck Citation2003) and PAUP version 4.0b10 (Sinauer Associates, Sunderland, MA) (Swofford Citation2002), respectively.
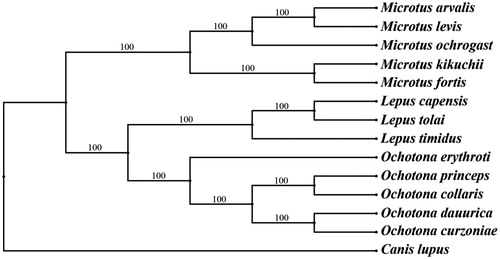
The different methods (BI and ML) obtained the same topology, with strong support for all nodes. The result of phylogenetic tree shows that O. dauurica was close to other Ochotona species, especially O. curzoniae (). Although O. dauurica looks like a mouse, it was actually more closely related to a rabbit, which was also supported by previous study (Wu Citation2003). We expect the data of this study to provide a useful for further research and phylogenetic relationship of Lagomorpha.
Figure 1. Bayesian phylogenetic inference (BI) and maximum-likelihood (ML) trees of 14 species based on 12 protein-coding genes except ND6 and the BI posterior probabilities are shown on the nodes. The species accession numbers were downloaded from GenBank are NC_038176 (M. arvalis), NC_008064 (M. levis), NC_027945 (M. ochrogaster), AF348082 (M. kikuchii), JF261174 (M. fortis), GU937113 (L. capensis), NC_025748 (L. tolai), KR019013 (L. timidus), NC_037186 (O. erythrotis), NC_005358 (O. princeps), AF348080 (O. collaris), EF535828 (O. curzoniae), KF857179 (C. lupus), respectively.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Additional information
Funding
References
- Lin YH, Waddell PJ, Penny D. 2002. Pika and vole mitochondrial genomes increase support for both rodent monophyly and glires. Gene. 294:119–129.
- Nylander JAA. 2004. MrModeltest version 2.1. Computer program distributed by the author. Uppsala, Sweden: Uppsala University.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Swofford DL. 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4.0b10. Sunderland (MA): Sinauer Associates.
- Wu S. 2003. The cranial morphology and phylogenetic relationship of Alloptox gobiensis (lagomorpha, ochotonidae). Vert Palas. 41:115–130.
- Zhong W, Wang G, Zhou Q, Wan X, Wang G. 2008. Effects of winter food availability on the abundance of Daurian pikas (Ochotona dauurica) in Inner Mongolian grasslands. J Arid Environ. 72:1383–1387.
