Abstract
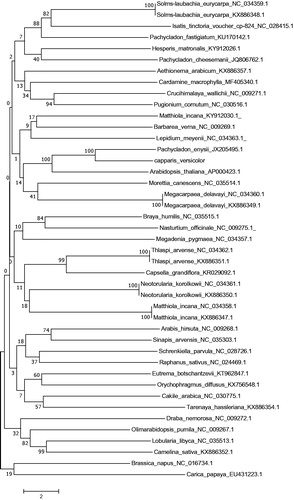
Capparis versicolor Griff. is a rare and threatened medicinal plant in South China. Chloroplast (cp)-derived markers have been widely used to study evolutionary relationships of many species. In this study, the complete cp genome of C. versicolor was assembled based on next-generation sequencing. The whole genome was 155,051 bp in length, consisting of a pair of inverted repeats, large single copy region, and a small single copy region. The cp genome contained 123 genes, including 83 protein-coding genes, 4 rRNA genes, and 36 trRNA genes. The overall GC content of the whole cp genome was 36.38%. A neighbour-joining phylogenetic analysis demonstrated that C. versicolor is clustered together with Pachycladon enysii.
Capparis versicolor Griff. is a perennial evergreen creeping shrub belonging to the genus flowering plants, which is mainly distributed in Guangxi and Guangdong province in China. It has used the mature seeds as a traditional Chinese medicine for pharyngitis (Satoru Nirasawa et al. Citation1994). Currently, C. versicolor has become one of rare and protected species in China due to habitat loss. On the other hand, the origin of the taxa is not accurately resolved and taxonomical position is still unclear. Hence, a better understanding of this species has a great potential in species discrimination and protection of species.
Fresh leaves of C. versicolor were collected from Yulin Normal University, Yulin city (Located at 109°39′ to 110°18′ east longitude, 22°19′ to 23°01′ north latitude), Guangxi Province, China. After morphological identification, the specimens were stored at −70 °C in our laboratory (Lab of Molecular Biology, Yulin Normal University). Genomic DNA was prepared in 350 bp paired-end libraries, tagged and subjected to the Illumina Hiseq 4000 (X-ten) sequencing system and yielded 3,116,666 Paired-End Raw Reads. Use SOAP denovo short sequence assembly software to assemble Clean Data to obtain optimal assembly results. GapCloser software repairs the inner hole of the assembly result and finally removes the redundant segment sequence to get the final assembly result. The circular C. versicolor chloroplast (cp) genome map was drawn using OGDraw version 1.2 (Lohse et al. Citation2007).
The C. versicolor cp genome is a covalently closed double-stranded circular molecule containing four segments of LSC, SSC, IRA, and IRB. The total length of sequence is 155,051 bp and the GC content is 36.38%. The cp genes were annotated using an online DOGMA tool (Wyman et al. Citation2004). A whole cp genome Blast (Altschul et al. Citation1990) search was performed against Kyoto encyclopedia of genes and genomes (Kanehisa Citation1997; Kanehisa et al. Citation2004, Citation2006) (KEGG), clusters of orthologous groups (Tatusov et al. Citation1997, Citation2003) (COG), non-redundant protein database (NR), Swiss-Prot (Magrane and UniProt Citation2011), and Gene Ontology (Ashburner et al. Citation2000) (GO), respectively. The C. versicolor cp genome consisted of 54.92% coding regions and 45.08% non-coding regions, including repeated sequences, transposons, and non-coding RNAs, and contained 123 unique genes, including 83 genes, 4 rRNA, and 36 tRNA.
To determine the evolutionary history of C. versicolor within the Rhoeadales, a neighbor-joining (NJ) phylogenetic tree was constructed with MEGA version 7 (Kumar et al. Citation2016). The result indicated that C. versicolor is clustered together with Pachycladon enysii what confirmed close relationship of Capparaceae and Brassicaceae). The complete cp genome information reported in this article will be a valuable resource for future studies on conservation genetics, taxonomy, phylogeny, and breeding of the Capparaceae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410.
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 25:25–29.
- Kanehisa M. 1997. A database for post-genome analysis. Trends Genet. 13:375–376.
- Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. 2006. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 34: D354.
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. 2004. The KEGG resource for deciphering the genome. Nucleic Acids Res. 32:D277–D280.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lohse M, Drechsel O, Bock R. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52:267–274.
- Magrane M, UniProt C. 2011. UniProt knowledgebase: a hub of integrated protein data. Database (Oxford). 2011:bar009.
- Satoru Nirasawa XL, Zhong H, Tomoko N, Masato K, Seiichi U, Yoshie K. 1994. Purification and structural characterization of homologues of the heat-stable sweet protein, mabinlin. Olfaction and Taste XI: Proceedings of the 11th International Symposium on Olfaction and Taste and of the 27th Japanese Symposium on Taste and Smell; July 12–16; Sapporo, Japan: Joint Meeting Held at Kosei-Nenkin Kaikan.
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 4:41.
- Tatusov RL, Koonin EV, Lipman DJ. 1997. A genomic perspective on protein families. Science. 278:631–637.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.