Abstract
We present the complete mitochondrial genome of the Epaulette Shark Hemiscyllium ocellatum, sequenced with 24 primer sets. The 16,728 bp long circular genome consisted of 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and a non-coding control region. Several protein-coding genes ended with incomplete stop codons (TA- or T–). Phylogenetic analysis indicated that H. ocellatum clusters within the Hemiscylliidae clade but separately from members of its sister genus Chiloscyllium. Utilization of the genome and primer sets presented here will be beneficial to future molecular studies involving H. ocellatum and other members of the Orectolobiformes.
The Epaulette Shark (Hemiscyllium ocellatum) is a small, benthic, oviparous shark inhabiting coral reef environments in north-eastern Australia (Allen et al. Citation2016). Molecular phylogeny studies involving this species are limited and often based on a single genetic marker (e.g. Naylor et al. Citation2012; Allen et al. Citation2013). As whole mitochondrial genomes can improve the resolution of elasmobranch phylogenies (Chen et al. Citation2013), we sequenced the complete mitochondrial genome of H. ocellatum to determine its phylogenetic placement within the Orectolobiformes.
DNA was extracted from a fin clip of an individual collected on Arlington Reef, Queensland (16°43′01″S; 146°01′58″ E) as outlined in Geraghty et al. (Citation2013). This sample (010 ES Skin) is currently stored at the Department of Biological Sciences, Macquarie University in 70% ethanol. Mitochondrial genome fragments were amplified using previously published and newly developed primer sets (see Supplementary Material available at https://doi.org/10.6084/m9.figshare.7300406). Forward and reverse reads of each fragment were aligned, trimmed, and checked for nucleotide assignment errors in Geneious® version 10.2.6 (http://www.geneious.com, Kearse et al. Citation2012). Edited fragments were mapped to the complete mitochondrial genome of Chilosyllium griseum (GenBank: JQ434458) to ensure complete coverage and then de novo assembled. The resulting sequence was annotated using the MitoAnnotator tool on the MitoFish website (Iwasaki et al. Citation2013), and checked with tRNAscan-SE (Lowe and Chan Citation2016) and by comparison with annotated hemiscyllid mitochondrial genomes from GenBank.
To assess the phylogenetic position of H. ocellatum, a maximum likelihood (ML) tree was generated in MEGA7 (Kumar et al. Citation2016). Ten complete mitochondrial genomes, consisting of eight orectolobiforms and two heterodontiforms, were downloaded from GenBank and MUSCLE aligned (Edgar Citation2004) with H. ocellatum. The ML tree was constructed with the most appropriate substitution model (GTR + I + G) as indicated by the corrected Akaike Information Criterion (AICc) in jModelTest2 (Guindon and Gascuel Citation2003; Darriba et al. Citation2012), 1000 bootstrap replications, the partial deletion setting, and all codon positions included.
The mitochondrial genome of H. ocellatum (GenBank: MK052932) was 16,728 bp in length and consisted of 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and a non-coding control region (D-loop). Base composition was: A (33%), C (24%), G (12.8%), and T (30.2%). The COI gene was the only PCG that started with a GTG codon; all other PCGs started with ATG. Several PCGs (ND2, COII, ND3, ND4, and Cyt-b) ended with an incomplete stop codon (TA- or T–), which are likely completed by post-transcriptional polyadenylation (Ojala et al. Citation1981). The control region was 1099 bp long.
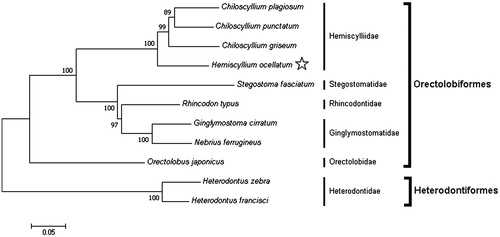
The ML tree () shows that H. ocellatum resides within the clade representing the family Hemiscylliidae. This phylogeny indicates that H. ocellatum does not cluster among members of the sister genus Chiloscyllium, as observed by Naylor et al. (Citation2012). Whole mitochondrial genome sequencing of other Hemiscyllium species and utilization of the genome presented here will help resolve the phylogenetic relationships within the genus. Additionally, the primer sets developed here are beneficial for future molecular studies involving both H. ocellatum and other sharks in the order Orectolobiformes.
Figure 1. Maximum likelihood tree showing the phylogenetic position of Hemiscyllium ocellatum (indicated by the star) within the order Orectolobiformes based on the complete mitochondrial genome. Members of the order Heterodontiformes served as the outgroup. Families are indicated by the vertical lines and orders by the square brackets. Scale bar indicates the number of substitutions per site and the numbers at the nodes indicate the percentage bootstrap values based on 1000 replications. GenBank Accession Numbers: Chiloscyllium plagiosum (FJ853422); C. punctatum (JQ082337); C. griseum (JQ434458); Stegostoma fasciatum (KU057952); Rhincodon typus (KF679782); Ginglymostoma cirratum (KU904394); Nebrius ferrugineus (KT852575); Orectolobus japonicus (KF111729); Heterodontus zebra (KC845548); H. francisci (NC_003137).
Supplemental Material
Download PDF (283.8 KB)Acknowledgements
The authors would like to thank James Cook University for providing the tissue sample and Macrogen (South Korea) for their Sanger sequencing service. We also thank L. Chow and T. Ghaly for their advice and technical assistance during this project.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Allen GR, Erdmann MV, Dudgeon CL. 2013. Hemiscyllium halmahera, a new species of bamboo shark (Hemiscylliidae) from Indonesia. Aqua. 19:123–136.
- Allen GR, Erdmann MV, White WT, Fahmi, Dudgeon CL. 2016. Review of the bamboo shark genus Hemiscyllium (Orectolobiformes: Hemiscyllidae). J Ocean Sci Found. 23:51–97.
- Chen X, Ai W, Ye L, Wang X, Lin C, Yang S. 2013. The complete mitochondrial genome of the grey bamboo shark (Chiloscyllium griseum) (Orectolobiformes: Hemiscylliidae): genomic characterization and phylogenetic application. Acta Oceanol Sin. 32:59–65.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797.
- Geraghty PT, Williamson JE, Macbeth WG, Wintner SP, Harry AV, Ovenden JR, Gillings MR. 2013. Population expansion and genetic structure in Carcharhinus brevipinna in the southern Indo-Pacific. Plos One. 8:e75169
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52:696–704.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30:2531–2540.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:W54–W57.
- Naylor GJP, Caira JN, Jensen K, Rosana KAM, Straube N, Lakner C. 2012. Elasmobranch phylogeny: a mitochondrial estimate based on 595 species. In: Carrier JC, Musick JA, Heithaus MR, editors. Biology of sharks and their relatives. 2nd ed. Boca Raton, FL: CRC Press; p. 31–56.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290:470–474.
