Abstract
The complete mitochondrial genome of Triplophysa sellaefer is a circular molecule of 16,571 bp in length with the A + T contents of 56.84%, containing 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and a putative control region. Six gene overlaps and 10 intergenic spacers were observed, with the total length of 26 bp and 34 bp, respectively. The phylogenetic analysis based on the concatenated PCG sequences revealed that T. sellaefer is closely related to T. alticeps. This study would provide useful information on phylogenetic and evolutionary studies in Triplophysa.
The genus Triplophysa is the largest group in Nemacheilinae subfamily with at least 140 valid species, and is mainly distributed in the Qinghai-Tibet Plateau (QTP) and its adjacent regions (Zhu Citation1989; Froese and Pauly Citation2017). In recent decades, the mitochondrial genomes of many Triplophysa species have been sequenced and used for phylogenetic and population genetics (Li et al. Citation2013; Chen et al. Citation2016; Jing et al. Citation2016; Wang et al. Citation2016). However, the information of mitogenome is lacking for several Triplophysa species that can be distributed in regions at lower altitudes and far from QTP. In this study, we firstly reported the complete mitochondrial genome of Triplophysa sellaefer which is mainly distributed in the middle and lower reaches of the Yellow River.
The specimens of T. sellaefer were collected from the Juma River (N 39°25′39.60″, E 115°9′55.34″), Baoding city, Hebei Province, China. After morphological identification, the specimens were stored in 100% alcohol at −20 °C. The total genomic DNA was extracted from a small piece of fin tissues using a traditional phenol–chloroform extraction method (Taggart et al. Citation1992), and is currently stored at Institute of Hydrobiology, Chinese Academy of Sciences. A set of primer pairs was designed and used for amplifying the mitochondrial genome sequences. The polymerase chain reaction was performed according to Feng et al. (Citation2018). After DNA sequencing, the complete mitochondrial genome of T. sellaefer was assembled and annotated, and has been deposited in GenBank database under the accession number KY851112.
The complete mitochondrial genome of T. sellaefer was 16,571 bp in length with the A + T contents of 56.84%, and contains 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes (12s rRNA and 16s rRNA), and a putative control region. Among these genes, the ND6 gene and eight tRNA genes were encoded on the L-strand, the remaining genes were encoded on the H-strand. Six gene overlaps and 10 intergenic spacers were observed, with the total length of 26 bp and 34 bp, respectively. The PCGs had the total length of 11,428 bp that encode 3800 amino acids. Most PCGs initiated with a common ATG start codon, while COI utilized GTG as start codon, which was similar to other Triplophysa fishes (Wang et al. Citation2016). Four types of the stop codons were observed in the PCGs, including TAA for COI, ATP8, ATP6, ND4L, ND5 and ND6, TAG for ND1, TA for ND4, and T for ND2, COII, COIII, ND3 and CYTB.
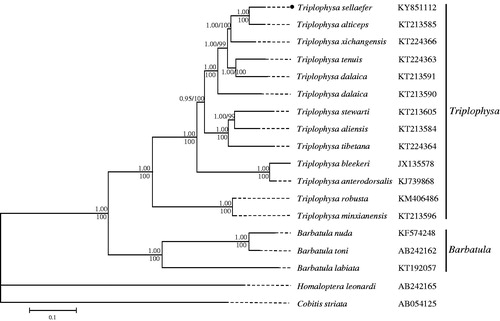
Based on the concatenated PCG sequences, the phylogenetic relationship among Triplophysa species was reconstructed by using the Bayesian inference (BI) and maximum-likelihood (ML) methods through MrBayes (Ronquist and Huelsenbeck Citation2003) and RAxML (Stamatakis Citation2006). Both the ML and BI phylogenetic trees showed an identical topology (). Within the genus Triplophysa, T. sellaefer is closely related to T. alticeps which is mainly distributed in the upper reaches of the Yellow River. All the Triplophysa species were clustered together and then grouped with three Barbatula species with high bootstrap values (99%), which was also reported in other studies (Tang et al. Citation2013; Liu and You Citation2016).
Figure 1. Bayesian inference and maximum likelihood (ML) phylogenetic trees based on 13 concatenated protein-coding genes. At each node of the left tree, the upper and lower numbers represent the Bayesian posterior probability and the bootstrap value for ML analyses, respectively. The GenBank accession number of each species was shown on the right of its name.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen S, Ya N, Xie C, Wang S, Ren D. 2016. Complete mitochondrial genome of the Triplophysa (Hedinichthys) yarkandensis (Day). Mitochondrial DNA B. 1:235–236.
- Feng X, He D, Sui X, Chen Y, Chen Y. 2018. Morphological and genetic divergence between lake and river populations of Triplophysa in Ngangtse Co, Tibet. Mitochondrial DNA Part A. 29:778–784.
- Froese R, Pauly D. 2017. FishBase. World Wide Web electronic publication. www.fishbase.org, version [2017 October].
- Jing H, Yan P, Li W, Li X, Song Z. 2016. The complete mitochondrial genome of Triplophysa lixianensis (Teleostei: Cypriniformes: Balitoridae) with phylogenetic consideration. Biochem Syst Ecol. 66:254–264.
- Li J, Si S, Guo R, Wang Y, Song Z. 2013. Complete mitochondrial genome of the stone loach, Triplophysa stoliczkae (Teleostei: Cypriniformes: Balitoridae). Mitochondrial DNA. 24:8–10.
- Liu T, You P. 2016. The complete mitochondrial genome of Triplophysa sp. (Teleostei: Cypriniformes: Balitoridae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:4557–4558.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Taggart JB, Hynes RA, Prodöuhl PA, Ferguson A. 1992. A simplified protocol for routine total DNA isolation from salmonid fishes. J Fish Biol. 40:963–965.
- Tang Q, Huang Y, Wang J, Huang J, Wang Z, Peng Z. 2013. The complete mitochondrial genome sequence of Triplophysa bleekeri (Teleostei, Balitoridae, Nemacheilinae). Mitochondrial DNA. 24:25–27.
- Wang Y, Shen Y, Feng C, Zhao K, Song Z, Zhang Y, Yang L, He S. 2016. Mitogenomic perspectives on the origin of Tibetan loaches and their adaptation to high altitude. Sci Rep. 6:29690
- Zhu SQ. 1989. The loaches of the subfamily Nemacheilinae in China (Cypriniformes: Cobitidae). Nanjing: Jiangsu Science and Technology Press; p. 68–132.
