Abstract
Arisaema ringens is a dioecious herb of the family Araceae. So far, there have been no studies on the genome of A. ringens. Here we report and characterize the complete plastid genome sequence of A. ringens in an effort to provide genomic resources useful for promoting its conservation. The complete plastome is 160,792 bp in length and contains the typical structure and gene content of angiosperm plastome, including two inverted repeat regions of 25,312 bp, a large single copy region of 88,915 bp, and a small single copy region of 21,253 bp. The plastome contains 114 genes, consisting of 80 unique protein-coding genes, 30 unique tRNA genes, and 4 unique rRNA genes. The overall A/T content in the plastome of A. ringens is 63.50%. The complete plastome sequence of A. ringens will provide a useful resource for the conservation genetics of this species as well as for the phylogenetic studies for Araceae.
Arisaema ringens (Thunb.) Schott is a plant of the family Araceae. It is a dioecious herb disjunct distributed in China (i.e. Zhejiang, Jiangsu, and Taiwan), Japan, and south Korea (Li et al. Citation2011; Fu et al. Citation2015). A. ringens has high ornamental value and medicinal value. It has the functions of eliminating swelling and dispersing phlegm, dispelling wind and dispelling wind shock and so on. So far, there have been no studies on the genome of A. ringens. Consequently, the genetic and genomic information is urgently needed to promote its systematics research and the development of conservation value of A. ringens. Here we report and characterize the complete plastid genome sequence of A. ringens (GenBank accession number: MK111107, this study) in an effort to provide genomic resources useful for promoting its conservation.
In this study, A. ringens was sampled from Ishima Island, Anan City, Tokushima Pref. Japan (33.85°N, 138.81°E). A voucher specimen (Wang et al. Ar8) was deposited in the Herbarium of the Institute of Tropical Agriculture and Forestry (HUTB), Hainan University, Haikou, China.
The experiment procedure is as reported in Zhu et al. (Citation2018). The plastome was annotated using Geneious R8.0.2 (Biomatters Ltd., Auckland, New Zealand) against the plastome of Colocasia esculenta (NC_016753.1) (Ahmed et al. Citation2012). The annotation was corrected with DOGMA (Wyman et al. Citation2004). A circular plastome map was generated using OGDRAW (http://ogdraw.mpimp-golm.mpg.de/) (Lohse et al. Citation2013).
The plastome of A. ringens was found to possess a total length 160,792 bp with the typical quadripartite structure of angiosperms, containing two inverted repeats (IRs) of 25,312 bp, a large single copy (LSC) region of 88,915 bp, and a small single copy (SSC) region of 21,253 bp. The plastome contains 114 genes, consisting of 80 unique protein-coding genes, 30 unique tRNA genes, and 4 unique rRNA genes. The overall A/T content in the plastome of A. ringens is 63.50%, which the corresponding value of the LSC, SSC, and IR region were 65.30%, 69.50%, and 56.70%, respectively.
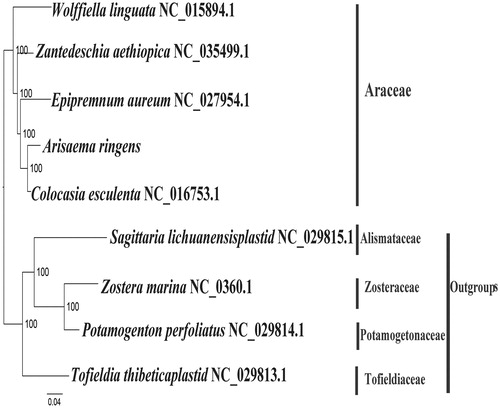
We used RAxML (Stamatakis Citation2006) with 1000 bootstraps under the GTRGAMMAI substitution model to reconstruct a maximum likelihood (ML) phylogeny of 5 published complete plastomes of Araceae, using Potamogeton perfoliatus, Sagittaria lichuanensis plastid, Tofieldia thibeticaplastid, and Zostera marina as outgroups. The phylogenetic analysis indicated that A. ringens iscloser to Colocasia esculenta (). Most nodes in the plastome ML trees were strongly supported. The complete plastome sequence of A. ringens will provide a useful resource for the conservation genetics of this species as well as for the phylogenetic studies for Araceae.
Figure 1. The best ML phylogeny recovered from nine complete plastome sequences by RAxML. Accession numbers: Arisaema ringens (Genbank, Accession number, MK111107, this study), Tofieldia thibeticaplastid NC_029813.1, Zantedeschia aethiopica NC_035499.1, Epipremnum aureum NC_027954.1, Colocasia esculenta NC_016753.1, Sagittaria lichuanensisplastid NC_029815.1. Outgroup: Potamogeton perfoliatus NC_029814.1, Zostera marina NC_036014.1, Wolffiella lingulata NC_015894.1.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Ahmed I, Biggs PJ, Matthews PJ, Collins LJ, Hendy MD, Lockhart PJ. 2012. Mutational dynamics of aroid chloroplast genomes. Genome Biol Evol. 4: 1316–1323.
- Fu Z, Ye L, Ye M, et al. 2015. The test on seeds breeding of rare flowers Arisaema ringens. Seed. 11:1001–4705.
- Li H, Guanghua Z, Peter CB, Jin M, Wilbert LAH, Josef B, Niels J. 2011. Araceae. In: Wu, ZY, Raven PH, Hong DY, editors. Flora of China. Vol.23. Beijing: Science Press.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. Organellar Genome-DRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zhu ZX, Mu WX, Wang JH, Zhang JR, Zhao KK, Friedman CR, Wang HF. 2018. Complete plastome sequence of Dracaena cambodiana (Asparagaceae): a species considered “Vulnerable” in Southeast Asia. Mitochondrial DNA B Resour. 3:620–621.
