Abstract
The complete mitochondrial genome of Trematomus pennellii was determined by MiSeq platform. The mitogenome of T. pennellii was 15,976 bp in length and encoded 37 genes (13 proteins, 22 tRNAs, 2 ribosomal RNAs) and a non-coding control region (D-Loop). Overall A + T contents (54.10%) in the mitogenome was higher than G + C contents (45.90%). Twenty-nine genes were located on H strand, whereas eight genes were identified on L strand. Among 13 protein-coding genes, unusual start codons were identified in COX1 (GTG), ND6 (ATT), COX2 (ATC), and ATP6 (ATC). The incomplete stop codon (T–) was identified in six genes including COXII, COXIII, NAD2, NAD3, NAD4, and Cytb. The phylogenetic tree of complete genome indicated that Trematomus pennellii was most closely related to Trematomus bernacchii with 91.65% DNA sequence identity.
As the unique single dominant teleost fish fauna in the Southern Ocean, genetic information of the Notothenioidei is important to understand its adaptive radiation in the freezing waters of the Southern Ocean. Genus Trematomus is a relatively young clade in the Notothenioidei, which had been evolved approximately 7.4 ± 0.3 million years ago and 15 species are currently reported (Near et al. Citation2004). Species in the genus are characterized by the possession of antifreeze glycoproteins and absence of hemoglobin in some species as the adaptive evolutionary mechanism (Kock Citation1992; Policansky Citation1994; Praebel et al. Citation2009; Eastman Citation2013). The sharp-spined notothenia, Trematomus pennellii is characterized by its brownish color with 2 or 3 large dark spots and 4–6 dorsal spines (Parkes Citation1992; Balushkin Citation2000). In this study, we determined the complete mitochondrial DNA sequence of T. pennellii, which is one of the important Trematomus species in Antarctica
Trematomus pennellii was collected on 13 February 2014 from the ocean near to the McMurdo Station and stored in the Korea Polar Research Institute (KOPRI), Republic of Korea. Its identification was confirmed from both morphological characteristics and COI sequence identity to GenBank database (GenBank number: HQ713272). Specimen and its DNA is stored in the Department of Marine Biology, Pukyong National University. Mitochondrial DNA was extracted by the mitochondrial DNA isolation kit (Abcam, UK) followed by the manufacturer’s protocol. Prior to a library construction, mitochondrial DNA was fragmented by Covaris M220 Focused-Ultrasonicator (Covaris Inc., San diego, CA). A library was prepared by TruSeq® RNA library preparation kit V2 (Illumina, San diego, CA) and sequencing was conducted by MiSeq sequencer (Illumina). Geneious® 11.0.2 software (Kearse et al. Citation2012) was used to assembly of raw reads to construct a complete mitogenome.
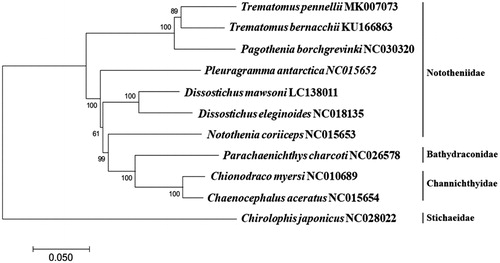
The complete mitochondrial genome of T. pennellii (GenBank Number: MK007073) was 15,976 bp in length, which consisted of 13 protein-coding genes, 22 tRNAs, 2 ribosomal RNAs (12S and 16S), and the non-coding control region (D-Loop). Overall A + T contents (54.10%) in the mitogenome was higher than G + C contents (45.90%). Twenty-nine and eight genes were located on the heavy (H) strand, and the light (L) strand, respectively. Nine protein-coding genes began with the typical start codon (ATG), while unusual ones also identified in COX1 (GTG), ND6 (ATT), COX2 (ATC) and ATP6 (ATC). Incomplete stop codon (T–) was identified in six genes including COXII, COXIII, NAD2, NAD3, NAD4, and Cytb. Except for tRNAser(GCT), all tRNAs were predicted to form a typical clover-leaf secondary structures according to ARWEN software (Laslett and Canbäck Citation2008). The phylogenetic tree of T. pennellii was constructed with nine complete mitogenomes in Notothenioids by MEGA7.0 program (Kumar et al. Citation2016) with minimum evolution (ME) algorithm. The result indicated that T. pennellii was most closely related to T. bernacchii among currently known species in Nototheniidae with 91.65% sequence identity (). More mitogenome sequences in Noththenioidei would help understand the unique evolutional relationship in teleost fish fauna in the Southern Ocean.
Figure 1. Phylogenetic tree of Trematomus pennellii in the suborder Notothenioidei. Phylogenetic relationship of Trematomus pennellii with its relatives was analyzed by MEGA7 software with Minimum Evolution (ME) algorithm and 1000 bootstrap replications. GenBank accession numbers were shown next to each scientific species name. The Chirolophis japonicus used as an outgroup species.
Disclosure statement
The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Balushkin A. 2000. Morphology, classification, and evolution of notothenioid fishes of the Southern Ocean (Notothenioidei, Perciformes). Journal Ichthyol. 40:S74.
- Eastman JT. 2013. Antarctic fish biology: evolution in a unique environment. San Diego, CA, USA: Academic Press Inc.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kock KH. 1992. Antarctic fish and fisheries. Cambridge, England: Cambridge University Press.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 33:1870–1874.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Near TJ, Pesavento JJ, Cheng C-HC. 2004. Phylogenetic investigations of Antarctic notothenioid fishes (Perciformes: Notothenioidei) using complete gene sequences of the mitochondrial encoded 16S rRNA. Mol Phylogenet Evol. 32:881–891.
- Parkes G. 1992. Fishes of the southern ocean. Rev Fish Biol Fisheries. 2:344–345.
- Policansky D. 1994. History and atlas of the fishes of the Antarctic Ocean. Science. 264:1002–1005.
- Praebel K, Hunt B, Hunt LH, DeVries AL. 2009. The presence and quantification of splenic ice in the McMurdo Sound notothenioid fish, Pagothenia borchgrevinki (Boulenger, 1902). Comp Biochem Physiol A Mol Integr Physiol. 154:564–569.
