Abstract
The complete mitochondrial genome of the morotoge shrimp, Pandalopsis japonica was determined by the high-throughput sequencing technique. The mitogenome of P. japonica (15,909 bp) encoded the 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes as well as the non-coding control region (D-Loop). The mitochondrial genome of P. japonica showed an identical gene arrangement in its mitochondrial genome to its relative Pandalid shrimps including P. borealis and P. hypsinotus. Among 13 compared mitogenomes within order Suborder Pleocyemata, P. japonica was most closely related to P. hypsinotus with 86% sequence identity clustering together with three other shrimps in family Pandalidae.
The morotoge shrimp, Pandalopsis japonica is mainly distributed in cold Sakhalin, East/Japan Sea, Pacific Ocean coast of Hokkaido, and Tohoku region (Komai and Mizushima Citation1993; Kim et al. Citation2006). It is important fishery resource in East/Japan Sea in Korea with its relatives, Pandalus hypsinotus and Lebbeus groenlandicus and its catches are conducted mainly on the seabed from 200 to 500 m in depth. Due to its high market price, the demands for the shrimp are increasing resulting in decreasing its native resource in Korea. In this report, the full mitochondrial genome sequence of P. japonica was determined by high-throughput sequencing technique, which would be used for the management of its resources.
P. japonica was collected near from Ulleungdo Island (37° 21′ 22N, 131° 27′ 06 E) and its identification was made by its morphological characteristics described in the Korean endemic species database (https://species.nibr.go.kr/index.do). Collected specimen and DNAs were stored at Department of Marine Biology in Pukyong National University, Korea. Purified mitochondrial DNA by a commercial kit (Abcam, UK) was fragmented into smaller sizes (∼350 bp) by Covaris M220 Focused-Ultrasonicator (Covaris Inc., Woburn, MA). A library for MiSeq sequencing was constructed by TruSeq® RNA library preparation kit V2 (Illumina, San Diego, CA). After the quantity of the library was confirmed by 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA), DNA sequencing was conducted by Illumina MiSeq sequencer (2 × 300 bp pair ends).
The complete mitochondrial genome of P. japonica was 15, 909 bp in length (GenBank Number; MK043326), which was similar to its relative, Pandalus borealis (Viker et al. Citation2006). This mitochondrial genome of P. japonica contained the highly conserved 13 protein-coding genes, 22 tRNA genes, and 2 rRNA (12S and 16S) as well as a non-coding control region (D-Loop). The overall gene arrangement of the P. japonica mitochondrial genome was identical to its relative Pandalus borealis in which eight genes were encoded on H strand (Viker et al. Citation2006). Unusual start codons were identified in six protein-coding genes including COX1, ATP8, ATP6, ND5, ND4, and ND2, while incomplete ‘T–’ was identified in COX2, ND3 and Cytb. Except for tRNA-Ser(TCT), all other 21 tRNA genes (63 bp to 73 bp) were predicted to form the typical secondary structures by ARWEN program (Laslett and Canbäck Citation2008).
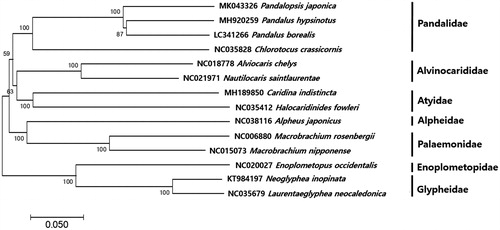
The phylogenetic position of P. japonica was analyzed by MEGA7 software with minimum evolutionary (ME) algorithm with 1000 bootstrap replicates (Kumar et al. Citation2016). Among13 compared mitogenomes in order Suborder Pleocyemata, P. japonica was clustered together with three other shrimps in family Pandalidae (). Within the family Pandalidae, P. japonica was most closely related to P. hypsinotus (86%) followed by P. borealis (85%) and Chlorotocus crassicornis (77%). The mitogenome information of P. japonica would contribute to its scientific conservation and management in Korea.
Disclosure statement
The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Kim DH, Choi JH, Kim JN, Cha HK, Oh TY, Kim DS, Han CH. 2006. Gonad and androgenic gland development in relation to sexual morphology in Pandalopsis japonica Balss, 1914 (Decapoda, Pandalidae). Crustaceana. 79:541–554.
- Komai T, Mizushima T. 1993. Advanced larval development of Pandalopsis Japonica Balss, 1914 (Decapoda, Garidea, Pandalidae) reared in the laboratory. Crustaceana. 64:24–39.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Viker SM, Klingberg ÅN, Sundberg P. 2006. The complete mitochondrial DNA sequence of the northern shrimp, Pandalus borealis. J Crustacean Biol. 26:433–435.