Abstract
The complete mitochondrial genome of spotted snakehead fish, Channa punctata (Bloch 1793) was determined by MiSeq platform. The genome was 16,409 bp in length, in which canonical 13 protein-coding genes, 22 tRNAs, 2 rRNAs, and a control region (D-Loop) were encoded. Relatively higher overall A + T contents (53.70%) was identified compared with G + C contents (46.30%). Among 13 protein-coding genes, only one unusual start condon (GTG) was identified in COX1, whereas six genes including COXII, COXIII, NAD2, NAD3, NAD4, and Cytb were ended with the incomplete stop codons (TA-/T–). Based on the currently identified mitogenomes of Channa fish species, C. punctata was most closely related to Channa gachua with 82% sequence identity.
The Spotted snakehead, Channa punctata (Bloch 1793) is a common air-breathing fish found in swamps, ditches, beels, and ponds (Jayaram Citation1981; Pethiyagoda Citation1991). This species widely distributed in the freshwaters and brackish water of Bangladesh, India, Pakistan, Afghanistan, Sri Lanka, Nepal, Myanmar, and China (Talwar and Jhingran Citation1991; Riede Citation2004; Mukhopadhyay and Ghosh Citation2007). Since C. punctata shares the habitat with its relatives such as Channa gachua, and Channa striata (Talwar and Jhingran Citation1991; Molur and Walker Citation1998; Froese and Pauly Citation2011), conservation and management of their resources based on the genetic information is strongly required.
C. punctata was collected from the river in Khulna, Bangladesh (22°50′44.3″ N, 89°32′27.7″E) in March 2017. The specimen was identified by its morphological characteristics and its COX1 sequence to the NCBI database (GenBank number: KT762386). The specimen is stored in the Department of Fisheries and Marine Science laboratory, Noakhali Science and Technology University, Noakhali, Bangladesh. The full mitochondrial DNA sequence of C. punctata was determined by next-generation sequencing (NGS) platform. Mitochondrial DNA was extracted by the mitochondrial DNA isolation kit (Abcam, UK) and the purified mitochondrial DNA was further fragmented into smaller sizes (∼350 bp) by Covaris M220 Focused-Ultrasonicator (Covaris Inc., Woburn, MA, USA). A library was constructed by TruSeq® RNA library preparation kit V2 (Illumina), and its quality and the quantity were analyzed by 2100 Bioanalyzer (Agilent Technologies). The high-throughput sequencing was performed by MiSeq platform with 600-cycle kit (Illumina). Geneious software (v.11.0.2) was applied for the mitogenome assembly (Kearse et al. Citation2012).
The complete mitochondrial genome of C. punctata (GenBank Number: MK007075) was 16,409 bp in length, which consisted of 13 protein-coding genes, 22 tRNAs, 2 ribosomal RNAs (12S and 16S), and a control region (D-Loop). Nine genes were encoded on the L strand while the other 28 genes were located on the H strand. Relatively higher overall A + T contents (53.70%) were identified compared with G + C contents (46.30%). Among 13 protein-coding genes, only one unusual start condon (GTG) was identified in COX1, whereas six genes including COXII, COXIII, NAD2, NAD3, NAD4, and Cytb were ended with the incomplete stop codons (TA-/T–).
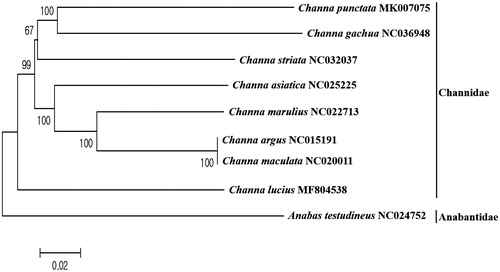
The phylogenetic tree was constructed to understand the evolutional relationship of C. punctata with other seven currently reported mitogenomes of Channa species using MEGA7.0 program with minimum evolutionary (ME) algorithm (Kumar et al. Citation2016). Based on the mitogenome sequences, C. punctata showed the highest similarity to C. gachua (GenBank number: NC036948) with 82% nucleotide sequence identity (). Based on the COX1 gene sequence, C. punctata was most closely related to C. gachua (MH156949) and C. striata (KX389278) with 84% sequence identity. Those result supported the previous result about the evolution of Asian snakehead fishes (Adamson et al. Citation2010) and more mitogenome sequences would help us to establish the clear evolutional relationship of snakehead fishes.
Figure 1. Phylogenetic tree of Channa punctata within Channidae family. Phylogenetic tree of Channa punctata complete genome was constructed by MEGA7 software with Minimum Evolution (ME) algorithm with 1000 bootstrap replications. GenBank Accession numbers were shown followed by each species scientific name. Anabas testudineus was used as an outgroup species.
Disclosure statement
The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Adamson EAS, Hurwood DA, Mather PB. 2010. A reappraisal of the evolution of Asian snakehead fishes (Pisces, Channidae) using molecular data from multiple genes and fossil calibration. Mol Phylogenet Evol. 56:707–717.
- Froese R, Pauly D. 2011. FishBase. 2011. World Wide Web electronic publication; [accessed 22 Feb 2011]. Available at: http://www/fishbase.org
- Jayaram KC. 1981. Freshwater Fishes of India, Pakistan, Bangladesh, Burma and Sri Lanka. agris.fao.org, In AGRIS,The Survey.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 33:1870–1874.
- Molur S, Walker S. 1998. Report of the Workshop on “Conservation assessment and management plan for freshwater fishes of India”. Zoo Outreach Organization, Convservation Breeding Specialist Group, Coimbatore, India.
- Mukhopadhyay T, Ghosh S. 2007. Lipid profile and fatty acid composition of two silurid fish eggs. J Oleo Sci. 56:399–403.
- Pethiyagoda R. 1991. Freshwater fishes of Sri Lanka. Wildlife Heritage Trust of Sri Lanka. Colombo, Sri Lanka, Wildlife Heritage Trust of Sri Lanka.
- Riede K. 2004. Global register of migratory species: from global to regional scales: final report of the R&D-Projekt 808 05 081. Bonn, Germany: Federal Agency for Nature Conservation.
- Talwar P, Jhingran A. 1991. Systematic account of Siluriformes fishes. In: Inland fishes of India and adjacent countries. Vol 2. A.A. Balkema: Rotterdam, Netherland; p.543–714.
