Abstract
The order Commelinales comprises many species with important economic and ecological values and represents a striking case of morphologically convergent evolution in monocots. However, so far the genomic resources of species within Commelinales are limited and the deep level phylogeny of Commelinales has not been well resolved. Here we reported and characterized the first complete chloroplast (cp) genome of Pontederia cordata and analysed the phylogenomic relationship of Commelinids based on complete cp sequences. The complete cp genome of P. cordata is 162,234 bp in length with a typical quadripartite structure. The overall GC content is 36.2%. A total of 124 genes were annotated, including 85 encoding genes, 38 transfer RNA genes, and 8 ribosomal RNA genes. The inverted repeat regions contain six CDSs, eight tRNA genes and all the four rRNA genes. Phylogenetic study validated the phylogenetic position of P cordata and showed that the three species from Commelinales form a monophyletic group sister to Zingiberales with high support rate. Meanwhile, species from Zingiberales and Commelinale form a molophyletic group sister to the Poales clade.
Pontederia cordata is a wetland plant species native to the North and South America which has now been widely cultivated in China for ornamental purposes. Since P. cordata can effectively inhibit the growth of algal, it is also considered as a promising plant for water environmental restoration currently (Qian et al. Citation2017). The order Commelinales represents a typical system of morphologically convergent evolution (Givnish et al. Citation1999). However, so far the deep level phylogeny of Commelinales has not been well resolved. To provide genomic resources for investigating the evolution of Commelinales, here we reported and characterized the complete chloroplast (cp) genome of P. cordata, the first plastome sequence in the Pontederiaceae family.
Leaf materials of P. cordata were collected on the Campus of Zhejiang University, Hangzhou city, Zhejiang, China. The corresponding specimen was deposited in the herbarium of Tarim University (TAU) (voucher number: Yang20180001). Total DNA was isolated using the DNA Plantzol Reagent (Invitrogen, Carlsbad, CA) and sent to BGI (Shenzhen, Guangdong, China) for next-generation sequencing on the Illumina Hiseq Platform (Illumina, San Diego, CA). The cp genome was assembled using NOVOPlasty (Dierckxsens et al. Citation2017) and annotated using Geneious version 11.0.4 (Biomatters Ltd., Auckland, New Zealand). The cp genome sequence of Xiphidium caeruleum (GenBank accession number: JX08669.1) was used as the reference (Barrett et al. Citation2013).
The cp genome of P. cordata is 162,234 bp in length with a typical quadripartite structure consisting of a large single copy of 90,112 bp, a small single copy region of 18,454 bp, and a pair of 26,834 bp inverted repeats (IRs). The GC content of the complete cp genome is 36.2%. The genome contains 124 genes, including 85 encoding genes, 38 transfer RNA genes, and 8 ribosomal RNA genes. The IR regions contain six coding sequences (CDSs), eight tRNA genes, and all the four rRNA genes. The cp genome structure, GC content, and gene locations of P. cordata are almost identical to the previously reported cp genomes of Xiphidium caeruleum and Hanguana malayana belonging to Commelinales (Barrett et al. Citation2013).
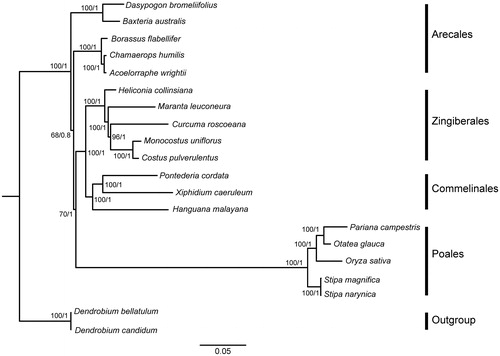
Phylogenomic analysis based on the complete cp genome was performed with the maximum likelihood (ML) and Bayesian inference (BI) methods (Ronquist and Huelsenbeck Citation2003; Stamatakis Citation2014) implemented on the CIPRES Science Gateway V.3.3 (Miller et al. Citation2010). Two species from Asparagales (Dendrobium bellatulum and Dendrobium candidum) were used as the outgroup. The cp genome sequences of P. cordata was submitted to GeneBank (accession number: xxxxx). The GenBank accession numbers of other species used for phylogenomic analysis are provided in . All the 20 genome sequences were aligned using MAFFT version 7.0 (Katoh et al. Citation2017).
Table 1. GenBank accession numbers of all the plastome sequences used for phylogenetic reconstruction.
The ML and BI analyses generated the same tree topology (). Pontederia cordata and two other species from Commelinales form a monophyletic group sister to Zingiberales. The evolutionary relationships of Poales, Arecales, and Asparagales are consistent with the results from previous phylogenetic study (Barrett et al. Citation2013). The P. cordata plastome reported in this study may provide useful resources for the development of ornamental and ecological value as well as robust phylogenetic study at deep level of Commelinales in the future.
Figure 1. Molecular phylogeny of Commelinids based on the complete chloroplast genomes of 20 species, with Dendrobium bellatulum and Dendrobium candidum as the outgroup. Relative branch lengths are indicated below the phylogenetic tree. Numbers above each branch indicates the bootstrap values obtained from ML analysis (before the slash) and the posterior probabilities obtained from BI analysis (after the slash).
Disclosure statement
The authors are really grateful to the opened raw genome data from public database. The authors report no conflict of interest and are responsible for the content and writing of the paper.
Additional information
Funding
References
- Barrett CF, Davis JI, Leebens-Mack J, Conran JG, Stevenson DW. 2013. Plastid genomes and deep relationships among the commelinid monocot angiosperms. Cladistics. 29:65–87.
- Dierckxsens N, Mardulyn P, Smits G. 2017. Novoplasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids. 45:e18. DOI: 10.1093/nar/gkw955.
- Givnish TJ, Evans TM, Pires JC, Sytsma KJ. 1999. Polyphyly and convergent morphological evolution in commelinales and commelinidae: evidence from rbc l sequence data. Mol Phylogenet Evol. 12:360–385.
- Katoh K, Rozewicki J, Yamada KD. 2017. Mafft online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 4:1-7. DOI: 10.1093/bib/bbx108.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the cipres science gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop. 14:1–8.
- Qian Y, Xu N, Liu J, Tian R. 2017. Inhibitory effects of pontederia cordata on the growth of microcystis aeruginosa. Water Sci Technol. 2017:99–107.
- Ronquist F, Huelsenbeck JP. 2003. Mrbayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Stamatakis A. 2014. Raxml version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
