Abstract
The complete mitochondrial genome in Mola carplet, Amblypharyngodon mola (Hamilton, 1822) was determined by MiSeq platform for the first time in the genus. The mitogenome of A. mola was circular and 16,545 bp in length, which harbours a canonical 13 protein-coding genes, 22 tRNAs, 2 rRNAs, and a control region (D-Loop). Twenty eight genes were encoded on the H strand and the other 9 genes were on the L strand. The incomplete stop codon (TA-/T–) was identified in seven genes (COX2, COX3, ND2, ND3, ND4, ATP6, and Cytb). The mitogenome of A. mola showed highest (82%) sequence identity with Rasbora vaterifloris (GenBank No. NC015531), which is endemic to Sri Lanka.
The Mola carplet, Amblypharyngodon mola (Hamilton, 1822) locally known as a mola or moia, is one of the most important freshwater small indigenous species (SIS) in Bangladesh. This species is used as the main source of animal protein and micronutrients for the rural people in Bangladesh, India, and the surrounding countries (Thompson et al. Citation2002; Kongsbak et al. Citation2008; Kunda et al. Citation2008). A. mola generally inhabits in ponds, canals and paddy fields of Bangladesh, India, Pakistan, and Myanmar (Jayaram Citation1981; Menon Citation1999). The habitat of this fish is currently threating by the various anthropogenic activities and conservation of the resources is required.
A. mola was collected from a pond in Khulna, Bangladesh (22°50'44.3" N, 89°32'27.7" E) in March 2017. The specimen was identified by morphological characteristics and COI sequence identity to the database (GenBank Number: KT364774). The specimen and its DNA are stored at Department of Fisheries and Marine Science laboratory, Noakhali Science and Technology University, Bangladesh, and Department of Marine Biology, Pukyong National University, Korea, respectively. Mitochondrial DNA was extracted by the mitochondrial DNA isolation kit (Abcam, Cambridge, UK) and the purified mitochondrial DNA was fragmented into smaller sizes (∼350 bp) by Covaris M220 Focused-Ultrasonicator (Covaris Inc., Woburn, MA). A library was constructed by TruSeq® RNA library preparation kit V2 (Illumina, San Diego, CA), and its quality and quantity were analysed by 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). The high-throughput sequencing was performed by MiSeq platform with 600-cycle kit (Illumina, San Diego, CA). Geneious software version 11.0.2 (Auckland, New Zealand) was applied for the mitogenome assembly (Kearse et al. Citation2012). Phylogenetic tree was constructed with full mitochondrial genomes using MEGA version 7.0 program with minimum evolutionary (ME) algorithm (Kumar et al. Citation2016).
The circular complete mitochondrial genome of A. mola (GenBank Number: MK070910) was 16,545 bp in length, which harbours 13 protein-coding genes, 22 tRNAs, two ribosomal RNAs (12S and 16S), and a control region (D-Loop). Compared with G + C contents, higher overall A + T contents (58.10%) were identified. Twenty eight genes were encoded on the H strand and the other 9 genes were on the L strand. Three out of 13 protein-coding genes (COX1, ND2, and ND4L) begin with the unusual start condon (GTG) and 7 of them (COX2, COX3, ND2, ND3, ND4, ATP6, and Cytb) end with the incomplete stop codon (TA-/T–).
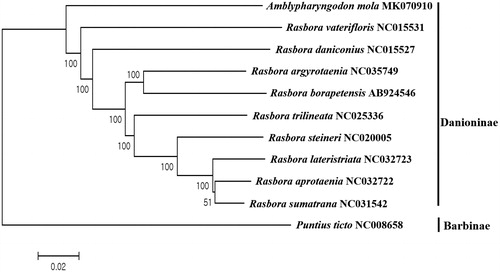
This is the first report about the mitogenome sequence in the genus Amblypharyngodon and the phylogenetic tree of A. mola was constructed with nine complete mitogenomes of its relatives in subfamily Danioninae. Based on the full mitogenome sequences to the NCBI database, A. mola showed the highest (82%) sequence identity with Rasbora vaterifloris (NC015531), which is the endemic freshwater species in Sri Lanka (). Based on the COI gene sequence, A. mola was most closely related to Amblypharyngodon melettinus (FJ751272) with 90% sequence identity, which is endemic to India and Sri Lanka. More mitogenome sequences would help us understand the evolutional relationship of fish in subfamily Danioninae in South Asia.
Figure 1. Phylogenetic tree of Amblypharyngodon mola within Danioninae Subfamily of Cyprinidae family. Phylogenetic tree of Amblypharyngodon mola complete genome was constructed by MEGA version 7 software by Minimum Evolution (ME) algorithm with 1000 bp replications. GenBank Accession numbers were shown followed by each species scientific name. Puntius ticto was used as an outgroup species.
Disclosure statement
The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Additional information
Funding
References
- Jayaram KC. 1981. Freshwater fishes of India, Pakistan, Bangladesh, Burma and Sri Lanka. Calcutta: The Survey.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kongsbak K, Thilsted SH, Wahed MA. 2008. Effect of consumption of the nutrient-dense, freshwater small fish Amblypharyngodon mola on biochemical indicators of vitamin A status in Bangladeshi children: a randomised, controlled study of efficacy. Br J Nutr. 99:581–597.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Kunda M, Azim ME, Wahab MA, Dewan S, Roos N, Thilsted SH. 2008. Potential of mixed culture of freshwater prawn (Macrobrachium rosenbergii) and self‐recruiting small species mola (Amblypharyngodon mola) in rotational rice–fish/prawn culture systems in Bangladesh. Aquacult Res. 39:506–517.
- Menon AGK. 1999. Check list–fresh water fishes of India. Vol. 175. Calcutta: The Survey.
- Thompson P, Roos N, Sultana P, Thilsted SH. 2002. Changing significance of inland fisheries for livelihoods and nutrition in Bangladesh. J Crop Prod. 6:249–317.
