Abstract
Plantago depressa Willd. (Plantaginaceae) belonging to the section Mesembrynia and subgenus Plantago has been used as a traditional medicine. Here, we presented the complete chloroplast genome of P. depressa which is 164,617 bp long and has four subregions: 82,933 bp of large single copy (LSC) and 4,908 bp of small single copy (SSC) regions are separated by 38,388 bp of inverted repeat (IR) regions including 139 genes (93 protein-coding genes, eight rRNAs, and 38 tRNAs). The overall GC contents of the chloroplast genome were 38.0% and in the LSC, SSC, and IR regions were 36.6%, 30.6%, and 39.9%, respectively. Phylogenetic tree constructed with concatenated 79 protein-coding genes sequences of the chloroplast genomes in Plantaginaceae from APG group presents the phylogenetic position of P. depressa.
Genus Plantago is a cosmopolitan genus and annual to perennial herbs. This genus with about 250 species is widely distributed in temperate regions. Several Plantago species has useful compounds and have been used as a traditional medicine from leaves and seeds including Plantago depressa Willd. (Pourmorad et al. Citation2006; Velasco-Lezama et al. Citation2006; Souri et al. Citation2008; Weryszko-Chmielewska et al. Citation2012; Han et al. Citation2016). Plantago depressa, belonging to the section Mesembrynia and subgenus Plantago (Ishikawa et al. Citation2009), is a perennial plant and usually grows in roads or fields in East Asia. Till now, there are two available complete chloroplast genomes, Plantago media in section Plantago of subgenus Plantago and Plantago maritima in section Maritima of subgenus Coronopus (Zhu et al. Citation2016).
Here, we sequenced complete chloroplast genome of P. depressa isolated in Republic of Korea (Sample was deposited in the InfoBoss Cyber Herbarium (IN); C-H. Park, IB-00570) as a representative chloroplast genome of the section Mesembrynia and subgenus Plantago. Total DNA of P. depressa was extracted from fresh leaves by using a DNeasy Plant Mini kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using HiSeq4000 at Macrogen Inc., Republic of Korea, and de novo assembly was conducted by Velvet 1.2.10 (Zerbino and Birney Citation2008), gap filling was done by SOAPGapCloser 1.12 (Zhao et al. Citation2011), and all bases were confirmed by alignment results generated by BWA 0.7.17 (Li Citation2013) and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.1.5 (Biomatters Ltd., Auckland, New Zealand) was used for chloroplast genome annotation based on P. media chloroplast genome (NC_028520; Zhu et al. Citation2016).
The chloroplast genome of P. depressa (Genbank accession is MK144833) is 164,617 bp and has four sub-regions: 82,933 bp of a large single copy (LSC) and 4,908 bp of small single copy (SSC) regions are separated by 38,388 bp of inverted repeat (IRs) regions. It contained 139 genes (93 protein-coding genes, eight rRNAs, and 38 tRNAs); 26 genes (15 protein-coding genes, 4 rRNAs, and 7 tRNAs) are duplicated in inverted repeat (IR) regions. The overall GC contents of P. depressa chloroplast genome were 38.0% and in the LSC, SSC, and IR regions were 36.6%, 30.6%, and 39.9%, respectively.
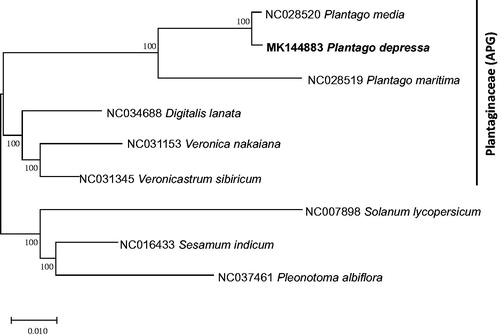
Concatenated alignment of 79 protein-coding genes from six chloroplast genomes in Plantaginaceae based on APG system (Chase et al. Citation2016) and three species from Pedaliaceae, Bignoniaceae, and Solanaceae, respectively, selected due to rapid evolution of Plantago chloroplast genomes (Zhu et al. Citation2016), were used for constructing neighbor joining phylogenetic tree using MEGA X (Kumar et al. Citation2018). Each multiple sequence alignment was conducted MAFFT 7.388 (Katoh and Standley Citation2013) after extracting the 79 protein-coding genes using at a pipeline in the Plant Chloroplast Database (PCD; in preparation). The phylogenetic tree confirmed that Plantago chloroplast has been evolved fast in comparison to other genera (). In addition, P. depressa is clustered into P. media even though two species are from different sections (). More Plantago chloroplast genomes from different subgenera will provide detailed dynamic evolutionary history to reconstruct the phylogenetic relationship of Plantago species.
Figure 1. Neighbor joining and Maximum parsimony phylogenetic tree (bootstrap repeat is 10,000) of six Plantaginaceae chloroplast genomes: Plantago depressa (MK144883; this study), Plantago media (NC_028520), Plantago maritima (NC_028519), Veronica nakaiana (NC_031153), Veronicastrum sibiricum (NC_031345), Pleonotoma albiflora (NC_037461), Digitalis lanata (NC_034688), Sesamum indicum (NC_016433), and Solanum lycoverscum (NC_007898). The numbers above branches indicate bootstrap support value of neighbor joining tree.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chase MW, Christenhusz M, Fay M, Byng J, Judd W, Soltis D, Mabberley D, Sennikov A, Soltis P, Stevens P. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181:1–20.
- Han N, Wang L, Song Z, Lin J, Ye C, Liu Z, Yin J. 2016. Optimization and antioxidant activity of polysaccharides from Plantago depressa. Int J Biol Macromol. 93:644–654.
- Ishikawa N, Yokoyama J, Tsukaya H. 2009. Molecular evidence of reticulate evolution in the subgenus Plantago (Plantaginaceae). Am J Botany. 96:1627–1635.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Pourmorad F, Hosseinimehr S, Shahabimajd N. 2006. Antioxidant activity, phenol and flavonoid contents of some selected Iranian medicinal plants. Afr J Biotechnol. 5(11):1142–1145.
- Souri E, Amin G, Farsam H, Barazandeh TM. 2008. Screening of antioxidant activity and phenolic content of 24 medicinal plant extracts.DARU. 16(2):83–87.
- Velasco-Lezama R, Tapia-Aguilar R, Román-Ramos R, Vega-Avila E, Pérez-Gutiérrez MS. 2006. Effect of Plantago major on cell proliferation in vitro. J Ethnopharmacol. 103:36–42.
- Weryszko-Chmielewska E, Matysik-Wozniak A, Sulborska A, Rejdak R. 2012. Commercially important properties of plants of the genus Plantago. Acta Agrobotanica. 65.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2.
- Zhu A, Guo W, Gupta S, Fan W, Mower JP. 2016. Evolutionary dynamics of the plastid inverted repeat: the effects of expansion, contraction, and loss on substitution rates. New Phytologist. 209:1747–1756.
