Abstract
Populus tomentosa narrow crown [P. tomentosa (NC)], natural distributes in northern China, is an economically important ecotype of P. tomentosa. In this study, we assembled its complete chloroplast (cp) genome. The total genome size of P. tomentosa (NC) is 156,368 bp in length, containing a large single copy region of 84,623 bp, a small single copy region of 16,533 bp, and a pair of inverted repeat regions of 27,606 bp. We annotated 130 genes in the genome, including 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes, 12 genes of which have 1 intron and 3 genes contain 2 introns. The overall GC content of the whole genome is 36.8%. Phylogenetic analysis using 24 complete cp genome of Salicaceae suggested that P. tomentosa (NC) is most closely related to P. alba. var. pyramidalis.
The Chinese white poplar (Populus tomentosa Carr.), as one of the main commercial hybrid species, is an indigenous species in northern China, which plays a vital role in ecological and environmental protection (Eckenwalder Citation1996; Zhu and Zhang Citation1997; Zhang et al. Citation2004; Zhang et al. Citation2006). Many wild P. tomentosa ecotypes have arisen during the evolution of the species (Zhang et al. Citation2007). Among them, P. tomentosa narrow crown [P. tomentosa (NC)] is one of the economically important ecotype of P. tomentosa. In addition, its morphological characteristics such as bark color, bark smoothness, and branches erect are remarkably similar to that of P. alba. var. pyramidalis. Knowledge of its genetic information would contribute to the study of genome diversity and species relationship. In this study, we assembled and characterized the complete chloroplast (cp) genome of P. tomentosa (NC) based on the Illumina pair-end sequencing data.
The samples of P. tomentosa (NC) were collected from Baoding city (Hebei, China; Coordinates: 115°47′E, 38°87′N; Altitude: 26 m). Total genomic DNA was isolated from fresh leaves of the P. tomentosa (NC) using the Ezup plant genomic DNA preps Kit (Sangon Biotech, Shanghai, China). The voucher specimens of P. tomentosa (NC) were deposited at the herbarium of Southwest Forestry University, and DNA samples were correctly stored at Key Laboratory of State Forestry Administration on Biodiversity Conservation in Southwest China, Southwest Forestry University, Kunming, China. Based on the known sequence of the Populus chloroplast genome, 33 primer pairs were designed and the whole cp genome of P. tomentosa (NC) were amplified using the LA-PCR with Takara PrimeSTAR GXL DNA polymerase followed the method described by Yang et al. (Citation2014) (Table S1). PCR products were sequenced by next generation sequenceing in Nextomics Biosciences. The annotation of the genome was performed using the program Geneious R8 (Biomatters Ltd, Auckland, New Zealand), and then manually confirmed by comparing with the cp genome of P. trichocarpa.
The plastome of P. tomentosa (NC) is a single circular molecule with the length of 156,368 bp. The complete cp genomes displayed the typical quadripartite structure, including a large single copy (LSC) region of 84,623 bp, a small single copy (SSC) region of 16,533 bp, and a pair of inverted repeats (IR) region of 27,606 bp. The overall GC content of the whole genome is 36.8%, while the corresponding values of the LSC (34.6%) and SSC (30.6%) are low than that of the IR regions (42.0%). This cp genome contains 130 functional genes, including 85 protein-coding genes, 37 tRNA genes, and 8 ribosomal RNA genes. Among them, 12 genes (atpF, ndhA, ndhB, petB, rpoC1, rpl2, trnK-UUU, trnG-UCC, trnL-UAA, trnV-UAC, trnI-GAU, and trnA-UGC) contain a single intron, and 3 genes (ycf3, rps12, and clpP) have 2 introns. There are 18 genes duplicated in the IR regions, of which 7 are protein-coding genes, 7 are tRNA, and 4 are rRNA genes.
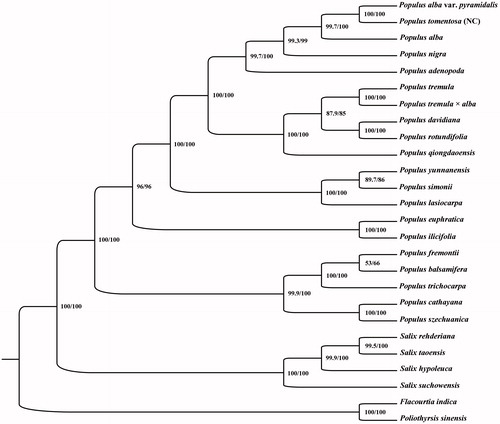
The phylogenetic relationship of P. tomentosa (NC) to other species in Salicaceae was evaluated by comparing the 24 species of Salicaceae chloroplast. Two species, Poliothyrsis sinensis and Flacourtia indica in the family of Flacourtiaceae, were used as out-group. The 26 complete cp sequences were aligned by the MAFFT version 7 software (Katoh and Standley Citation2013) Phylogenetic analysis was conducted base on maximum likelihood (ML) analyses were implemented in IQ-TREE 1.5.5 (Nguyen et al. Citation2015) under the GTR + F+R2 nucleotide substitution model, which was selected by ModelFinder (Kalyaanamoorthy et al. Citation2017). Support for the inferred ML tree was inferred by bootstrapping with 1000 replicates (Gao et al. Citation2018). The phylogenetic analysis showed that P. tomentosa (NC) appeared to be closely related to P. alba. var. pyramidalis ().
Figure 1. Phylogenetic relationships among 24 complete chloroplast genomes of Salicaceae. Bootstrap support values are given at the nodes. Chloroplast genome accession number used in this phylogeny analysis: P. adenopoda: KX425622; P.alba: AP008956; P. alba. var. pyramidalis: MG262344; P. balsamifera: KJ664927; P. cathayana: KP729175; P. davidiana: KX306825; P. euphratica:KJ624919; P. fremontii: KJ664926; P. ilicifolia: KX421095; P. lasiocarpa: KX641589; P. nigra: KX377117; P. qiongdaoensis: KX534066; P. rotundifolia: KX425853; P. simonii: MG262356; P. tomentosa narrow crown: MK252102; P. tremula: KP861984; P. tremula × alba: KT780870; P. szechuanica: MG262357; P. trichocarpa: EF489041; P. yunnanensis: KP729176; S. hypoleuca: MG262363; S. rehderiana: MG262367; S. suchowensis: KM983390; S. taoensis: MG262369; Flacourtia indica: MG262341; Poliothyrsis sinensis: MG262343.
Data Archiving Statement
The chloroplast genome data of the Populus tomentosa narrow crown will be submitted to Genebank of NCBI through the revision process. The accession numbers from Genebank must be supplied before the final acceptance of the manuscript.
Supplemental Material
Download MS Word (15.9 KB)Disclosure statement
The authors report no conflict of interest.
Additional information
Funding
References
- Eckenwalder JE. 1996. Systematics and evolution of Populus. In: Stettler RF, Bradshaw HD, Heilman JPE, Hinckley TM, editors. Biology of Populus and its implications for management and conservation. Ottawa (ON): NRC Research Press; p. 7–32.
- Gao FL, Shen JG, Liao FR, Cai W, Lin SQ, Yang HK, et al. 2018. The first complete genome sequence of narcissus latent virus from Narcissus. Arch Virol. 163:1–4.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14:587–589.
- Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs.Mol EcolResour.14:1024–1031.
- Zhang D, Zhang Z, Yang K, Li B. 2004. Genetic mapping in (Populus tomentosa x Populus bolleana) and P. tomentosa Carr. using AFLP markers. Theor Appl Genet. 108:657–662.
- Zhang DQ, Zhang ZY, Yang K. 2007. Identification of AFLP markers associated with embryonic root development in Populus tomentosa. Silvae Genet. 56:27–32.
- Zhu ZT, Zhang ZY. 1997. The status and advances of genetic improvement of Populus tomentosa Carr. Beijing Forestry Univ. 6:1–7.
- Zhang D, Zhang Z, Yang K. 2006. QTL analysis of growth and wood chemical content traits in an interspecific backcross family of white poplar (Populus tomentosa × P. bolleana) × P. tomentosa. Can. J. For. Res. 36:2015–2023.
