Abstract
Lovenula raynerae is the largest known African freshwater copepod. To date, it has only been sampled from ephemeral freshwater ecosystems. This paper reports the complete mitochondrial genome of L. raynerae, which was found to be 14,365 bp long. A base composition of 33.5% base A, 19.3% base G, 34.6% base T, and 12.5% base C was found, with 13 protein-coding genes, 22 tRNAs, and 2 rRNAs. This paper contributes to an improved understanding of phylogenetic relationships in an important crustacean group.
Copepods are a diverse group of aquatic crustaceans found in both marine and freshwater environments (Boxshall and Defaye Citation2008; Kim et al. Citation2013; Battuello et al. Citation2017). Of the 10 copepod orders (Boxshall and Defaye Citation2008), Calanoida is particularly important, as it is both species-rich (∼2266 species have been described; Battuello et al. Citation2017) and very abundant (they may constitute up to 95% of marine plankton samples; Mauchline Citation1998) and thus plays an important role in trophic interactions as a link between primary producers and secondary consumers (Dalu et al. Citation2016; Battuello et al. Citation2017; Wasserman et al. Citation2018). Despite their ecological importance and abundance, only one complete mitogenome has been published for the order Calanoida (Kim et al. Citation2013).
In this study, we describe the second complete mitochondrial genome of a calanoid copepod, that of Lovenula raynerae (Suarez-Morales et al. 2015). This recently described freshwater copepod has so far been found exclusively in ephemeral ponds in the Eastern Cape of South Africa (Suarez-Morales et al. 2015). One individual was collected in an ephemeral pond near Grahamstown, South Africa (33.250705°S, 26.436940°E). DNA was extracted using the CTAB method (Doyle and Doyle Citation1987) and the sample was then sequenced as follows. The gDNA was sonicated to ∼500 bp fragment size using the Covaris® Ultrasonicator (Covaris, Woburn, MA) and processed using the NEBNext Ultra DNA library prep kit for Illumina (New England Biolabs, Ipswich, MA). The library was subsequently denatured and sequenced on a MiSeq desktop sequencer (Illumina, San Diego, CA) at Monash University Malaysia. Mitogenome assembly was performed with MITObim version 1.8 (Hahn et al. Citation2013) using the 16S rRNA sequence of Mastigaodiaptomus nesus (Accession Number: EU582541.1) as the initial bait template. The mitogenome was manually re-circularized and re-oriented to the COI gene prior to submission to MITOS (Bernt et al. Citation2013) for annotation.
A sequence with a total length of 14,365 bp was generated whose base composition was 33.5% base A, 19.3% base G, 34.6% base T, and 12.5% base C. The sequence contained 13 protein-coding genes, 22 tRNAs and 2 rRNAs.
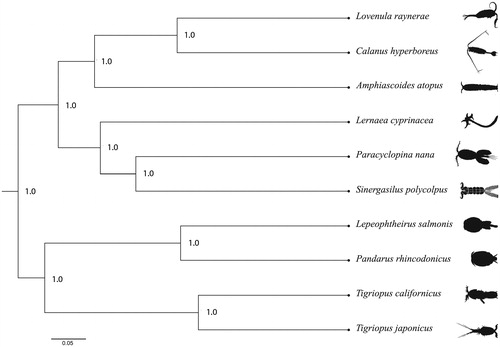
A phylogenetic tree was reconstructed using all 9 published mitogenomes from the subclass Copepoda. Due to extensive gene reshuffling in this group (Minxiao et al. Citation2011) the sequences were separated into the protein-coding genes, which were aligned separately via MAFFT (Katoh et al. Citation2017). A Bayesian phylogenetic tree was constructed using an RB substitution model (Bouckaert et al. Citation2013; Drummond and Bouckaert Citation2015) in BEAST v.2.5.0 (Bouckaert et al. Citation2014) with a chain length of one billion and a burn-in of 25%. The resulting tree was visualized in Figtree v.1.4.3 (Rambaut Citation2016). The phylogenetic tree () shows that L. raynerae is monophyletic with Calanus hyperboreus and that these two calanoid copepods have a sister taxon relationship with Amphiascoides atopus.
Figure 1. Bayesian phylogenetic tree of Lovenula raynerae and 9 other copepods. The numbers next to the nodes are posterior probabilities. Accession numbers are as follows: MH_710604 Lovenula raynerae, NC_019627 Calanus hyperboreus, NC_023783 Amphiascoides atopus, NC_025239 Lernaea cyprinacea, NC_012455 Paracyclopina nana, NC_028085 Sinergasilus polycolpus, NC_007215 Lepeophtheirus salmonis, NC_024046 Pandarus rhincodonicus, NC_008831 Tigriopus californicus, NC_003979 Tigriopus japonicus.
Acknowledgements
The authors are grateful to the Centre for High-Performance Computing (CHPC) in Cape Town for bioinformatics support
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Battuello M, Sartor RM, Brizio P, Nurra N, Pessani D, Abete MC, Squadrone S. 2017. The influence of feeding strategies on trace element bioaccumulation in copepods (Calanoida). Ecol Indic. 74:311–320.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Bouckaert R, Alvarado-Mora MV, Rebello Pinho JR. 2013. Evolutionary rates and HBV: issues of rate estimation with Bayesian molecular methods. Antivir Ther. 18:497–503.
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: a software platform for bayesian evolutionary analysis. PLoS Comput Biol. 10:e1003537.
- Boxshall GA, Defaye D. 2008. Global diversity of copepods (Crustacea: Copepoda) in freshwater. Hydrobiologia. 595:195–207.
- Dalu T, Weyl OLF, Froneman PW, Wasserman RJ. 2016. Trophic interactions in an austral temperate ephemeral pond inferred using stable isotope analysis. Hydrobiologia. 768:81–94.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Drummond AJ, Bouckaert RR. 2015. Bayesian evolutionary analysis with BEAST. Cambridge (UK): Cambridge University Press.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads - a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Katoh K, Rozewicki J, Yamada KD. 2017. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. Available from: http://dx.doi.org/10.1093/bib/bbx108
- Kim S, Lim B-J, Min G-S, Choi H-G. 2013. The complete mitochondrial genome of Arctic Calanus hyperboreus (Copepoda, Calanoida) reveals characteristic patterns in calanoid mitochondrial genome. Gene. 520:64–72.
- Mauchline J. 1998. The biology of Calanoid copepods. In: Blaxter J, Douglas B, Tyler P, editors. Advances in marine biology. 33rd ed. London: Academic Press; p. 49-97. ISBN: 978-0-12-105545-5.
- Minxiao W, Song S, Chaolun L, Xin S. 2011. Distinctive mitochondrial genome of Calanoid copepod Calanus sinicus with multiple large non-coding regions and reshuffled gene order: useful molecular markers for phylogenetic and population studies. BMC Genomics. 12:73.
- Suarez-Morales E, Wasserman R, Dalu T. 2015. A new species of Lovenula Schmeil (Copepoda, Calanoida, Diaptomidae) from the Eastern Cape Province of South Africa. Crustaceana. 88:324–342.
- Rambaut A. 2016. Figtree 1.4.3. https://doi.org/10.1371/journal.pcbi.1003537 [Last accessed 2018 Jun 06].
- Wasserman RJ, Weston M, Weyl OLF, Froneman PW, Welch RJ, Vink TJF, Dalu T. 2018. Sacrificial males: the potential role of copulation and predation in contributing to copepod sex-skewed ratios. Oikos.127:970-980.
