Abstract
Fengjing pig is one of the famous local breeds in China. In this study, we reported the complete mitochondrial genome of the Fengjing pig. The complete mitogenome sequence is 16,720 bp in length, containing the typical complement of 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNAs, and a non-coding control region. The new sequenced complete mitogenome of the Fengjing pig will provide valuable information for the application to conservation genetics and evolutionary studies for Chinese local pig breeds.
Fengjing pig is one of the excellent local breeds for its unique characteristics of higher reproductive performance, better meat quality and stronger ability of coarse feed like other Taihu pig breeds. Ear tissue samples were collected from Fengjing Pig Conservation Breeding Farm in Jurong City of Jiangsu Province (32°01′ 33.21′′ latitude, 119°14′ 48.92′′ longitude). The complete mitochondrial genome of Fengjing pig was amplified using polymerase chain reaction with 24 pairs of primers designed according to mitogenome of the Meishan pig (GenBank Accession No. KM998967.1) (Dong et al. Citation2016). DNA sequence was analysed by using tRNAscan-SE on-line (Lowe and Chan Citation2016) and MEGA X software (Kumar et al. Citation2018). Mitogenome sequence was deposited to NCBI through Blankit (GenBank Accession No. MH603005).
The complete mtDNA of Fengjing pig is 16,720 bp in length and the base nucleotide composition is 34.7% A, 25.8% T, 26.2% C, and 13.3% G, with an A + T (60.5%)-rich feature. It contains 2 ribosomal RNA genes (12S rRNA and 16S rRNA), 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNA), and a non-coding control region (D-loop). The distribution of these genes was the same as other reported pig breeds. Most protein initiation codons are ATG. However, ND2, ND3, and ND5 are ATA, and ND4L is GTG. And the terminate codons are TAG (ND1, ND2), TAA (COX1, ATP8, ATP6, ND4L, ND5, and ND6), GTG (CYTB) and an incomplete termination codon T (COX2, COX3, ND3, and ND4). The presumed 22 tRNA genes were distributed throughout the whole genome, ranging from 59 to 75 bp in size. The sequence length of 12s rRNA and 16s rRNA is 961 and 1570 bp, D-loop is 1284 bp containing 25 repeating units of 10 bp (CGTGCGTACA). The genes in the mitochondrial genome were arranged closely and total length of 8 overlaps was 76 bp, the largest overlap between ATP8 and ATP6 is 43 bp. The total length of 11 intergenic intervals is 57 bp. Twenty-nine genes and D-loop are located in the heavy chain, except for the PCG (ND6) and 8 tRNA genes (tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser, tRNA-Glu, and tRNA-Pro) are encoded in the light chain.
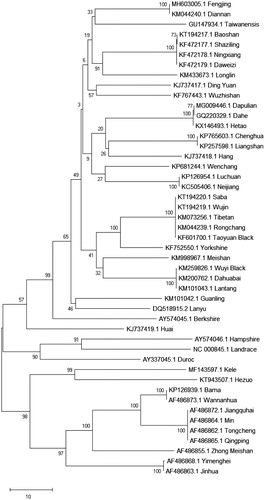
To assess the genetic relationship among different pig breeds in China, phylogenetic analysis was performed using the complete mitochondrial DNA sequences of 47 pig breeds. Each of the sequence datasets was aligned by clustal W in MEGA X and analysed by neighbour-joining (N-J) in MEGA X with 1000 bp replicates. The N-J tree showed that Chinese pig genetic resources were abundant and presented cluster distribution (). Fengjing pig is closely related to Diannan pig, which may be different from the traditional regional classification, and needs to study further. The three foreign breeds of Hampshire, Landrace, and Duroc are far apart from Chinese cluster distribution. This may be explained by the domestication of pigs from various regions of the world, which exhibited an independent origin of domestic pigs in Asia and Europe (Giuffra et al. Citation2000; Kijas and Andersson Citation2001; Larson et al. Citation2005).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Additional information
Funding
References
- Dong L, Ran M, Li Z, Chen B. 2016. The complete mitochondrial genome sequence of Meishan pig (Sus Scrofa) and a phylogenetic study. Mitochondrial DNA Part B. 1:112–113.
- Giuffra E, Kijas JM, Amarger V, Carlborg O, Jeon JT, Andersson L. 2000. The origin of the domestic pig: independent domestication and subsequent introgression. Genetics. 154:1785–1791.
- Kijas JM, Andersson L. 2001. A phylogenetic study of the origin of the domestic pig estimated from the near-complete mtDNA genome. J Mol Evol. 52:302–308.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Larson G, Dobney K, Albarella U, Fang M, Matisoo-Smith E, Robins J, Lowden S, Finlayson H, Brand T, Willerslev E, et al. 2005. Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science. 307:1618–1621.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:W54–W57.