Abstract
Isoetes yunguiensis is a rare and endangered fern endemic to Yunnan–Guizhou Plateau, China. The complete chloroplast genome sequence of I. yunguiensis was generated by de novo assembly using whole genome next-generation sequencing. The cp genome of I. yunguiensis was a circular structure and 145,355 bp in length, comprising a pair of inverted repeat (IR) regions of 26,130 bp, a large single copy (LSC) of 91,923 bp, and a small single copy (SSC) of 27,302 bp. The genome encoded 124 unique genes consisting of 82 protein-coding genes, 32 transfer RNA, four ribosomal RNA genes, and six pseudogenes. A maximum-likelihood phylogenetic analysis revealed that I. yunguiensis was closely related to I. butleri, I. engelmannii, I. flaccida, I. melanospora, and I. valida, with strong support values.
Isoetes yunguiensis Wang & Taylor (Isoetaceae) is an endemic fern to Yunnan–Guizhou Plateau, China (Wang et al. Citation2002; Zhang and Taylor Citation2013). It is similar to I. japonica Braun in its appearance and emergent aquatic habit, but differs from I. japonica in megaspore, microspore morphology, and chromosome number (2n = 22). At present, I. yunguiensis was recorded at few sites with less more 5,000 plants (Wang et al. Citation2002) and listed as critically endangered (CR) (Qin et al. Citation2017). Here, we assembled the complete chloroplast genome of I. yunguiensis to provide genomic resources for conservation and breeding of the taxon.
The mature leaves of I. yunguiensis were collected from Dapingqing National Wetland Park, Nayong County, Guizhou Province, China (105°27′32′′N and 26°40′39′′E) and preserved them in liquid nitrogen for further study. Voucher specimens (NY2016081206) were deposited at BJ (Bijie University Herbarium, Bijie City, Guizhou Province, China). The isolated genomic DNA was manufactured to average 150 bp paired-end (PE) library using the Illumina Hiseq platform, and sequenced by Illumina genome analyser. Chloroplast genome-related reads were sieved by mapping to the published chloroplast genome sequences in Isoetaceae. All related reads were assembled to contigs using SOAPdenovo2 (Luo et al. Citation2012), and all contigs were sorted and joined into a single-draft sequence using Geneious version 11.0.4 (Kearse et al. Citation2012) by comparison with the chloroplast sequence of I. valida (NC-038074). The draft sequence was then corrected manually by clean read mapping using bowtie2 (Langmead and Salzberg Citation2012) and Tablet (Milne et al. Citation2013). The genes in chloroplast genome were predicted using Geneious version 11.0.4 and corrected manually.
The complete chloroplast genome of I. yunguiensis (GenBank accession MK047605) was a circular form of 145,355 bp in length, comprising a large single copy (LSC) region of 91,923 bp and a small single copy (SSC) region of 27,302 bp, separated by a pair of 26,130 bp inverted repeat (IR) regions. The overall GC contents of I. yunguiensis cp genome was 38%, and while it was 48% in IR regions, which was higher than LSC (36.5%) and SSC (33.4%) regions. The genome contained 124 unique genes, including 82 protein-coding genes, 32 transfer RNA, four ribosomal RNA genes, and six pseudogenes. The genes accD, infA, rps2, rps16, tufA, an extra fragment of ndhB were interpreted as pseudogenes in our study. These pseudogenes were also found in I. flaccida (Karol et al. Citation2010). Among the annotated genes, 15 genes had a single intron and two genes harboured two introns.
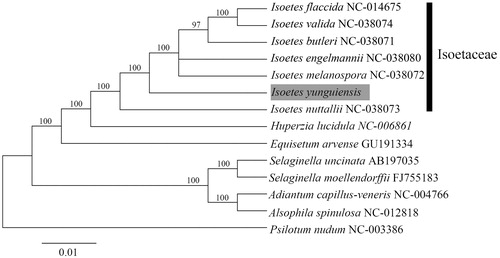
We compared the phylogenetic relationship of I. yunguiensis with other 13 species from the Isoetes using sequences obtained from GenBank. The phylogenetic tree was constructed using RAxML under the GTR + G nucleotide substitution model and 1000 rapid bootstraps (Stamatakis Citation2014). The ML tree indicated that Isoetes is monophyletic and I. yunguiensis forms a sister relationship with I. butleri, I. engelmannii, I. flaccida, I. melanospora, and I. valida with 100% bootstrap support values (). The chloroplast resource may be utilized for conservation genetics and breeding of I. yunguiensis.
Acknowledgements
We are grateful to the editor and anonymous reviewers for providing valuable comments on the manuscript. We thank Dr. Yi Qi Deng, Deng Feng Xie, MA. Hai Ying Liu for their help in preparing this article.
Disclosure statement
The authors report no conflicts of interest.
Additional information
Funding
References
- Karol KG, Arumuganathan K, Boore JL, Duffy AM, Everett KD, Hall JD, Kellon Hansen S, Kuehl JV, Mandoli DF, Mishler BD, et al. 2010. Complete plastome sequences of Equisetum arvense and Isoetes flaccida: implications for phylogeny and plastid genome evolution of early land plant lineages. BMC Evol Biol. 10:321.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–U354.
- Luo RB, Liu BH, Xie YL, Li ZY, Huang WH, Yuan JY, He GZ, Chen YX, Pan Q, Liu YJ, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 1:18.
- Milne I, Stephen G, Bayer M, Cock PJA, Pritchard L, Cardle L, Shaw PD, Marshall D. 2013. Using tablet for visual exploration of second-generation sequencing data. Brief Bioinform. 14:193–202.
- Qin H, Yang Y, Dong S, He Q, Jia Y, Zhao L, Yu S, Liu H, Liu B, Yan Y, et al. 2017. Threatened species list of China’s higher plants. Biodivers Sci. 25:696–744.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wang QF, Liu X, Taylor WC, He ZR. 2002. Isoetes yunguiensis (Isoetaceae), a new basic diploid quillwort from China. Novon. 12:587–591.
- Zhang LB, Taylor WC. 2013. Isoetaceae. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 2–3. St. Louis (MO): Science Press, Beijing and Missouri Botanical Garden Press; p. 35–36.