Abstract
In the present report, we describe the complete mitochonadrial genome of Nassarius hepaticus. The genome is found to be 15732 bp in length, which contained the typical mitogenome gene set of 22 tRNAs, 13 protein coding genes and 2 rRNAs. 13 protein-coding genes started with ATG; the stop codon in each of these protein-coding genes was either TAA or TAG, whereas NAD2 ends with AGT. The heavy strand was predicted to contain 13 protein-coding genes, 2 rRNA genes, 2 ATP synthase genes and 13 tRNA genes. The light strand was predicted to contain 8 tRNA. The phylogenetic tree was conducted with other 8 species with high similarity based on the method of maximum likelihood, the result could be a useful basis for management and utilization of this species.
Nassarius hepaticus, with a benthic life style, belongs to the genus of Nassarius and is distributed throughout worldwide oceans (Cernohorsky Citation1972), also be distributed in southeast coastal areas and other areas in China. Reports on N. hepaticus mainly focus on toxicity, the COI gene sequences, and phylogeny. Even if the classification on N. hepatic has long been controversial (Bargues and Mas-Coma Citation1997; Wang et al. Citation2007), however, only few reliable genetic studies related to N. hepaticus phylogenetics (Passamaneck et al. Citation2004; Li et al. Citation2010). In order to provide useful information for the future research of genetic diversity and phylogenetics, we determined the complete mitochondrial genome of N. hepaticus.
The specimens were collected from the north coastal water of Zhejiang (29°32′–31°04′N; 121°30′–123°25′E) and stored in national engineering research centre for marine aquaculture museum, we sequenced the whole mitochondrial genome of N. hepaticus using the Illumina HiSeq 2000 platform (Genome Analyser, Illumina, San Diego, CA). The annotated mitogenome of N. hepaticus has been deposited into GenBank database (GenBank accession number MH885313).
The N. hepaticus mitochondrial genome forms a 15,732 bp closed loop. The mitochondrial genome represents a typical Gastropoda mitochondrial genome in which it comprises 13 protein-coding genes, 22 putative tRNAs, and two rRNA genes, the average GC content of the N. hepaticus mitochondrial genome was 44.59%, which was much lower than the AT content, this feature was similar to the typical mitogenome of other invertebrate. Overall, 13 protein-coding genes started with ATG, these starting codons have been found to be common in the most of Gastropoda mitochondrial genome. The stop codon in each of these protein-coding genes was either TAA or TAG, whereas NAD2 ends with AGT. Most of the genes are encoded on the heavy strand (H-strand), the heavy strand was predicted to contain 13 protein-coding genes, including NADH dehydrogenase genes, cytochrome c oxidase genes, and ubichinol cytochrome c reductase genes (NAD1-6, NAD4L, COXI-III, Cob) and 2 ATP synthase genes (ATP6 and ATP8). Meanwhile, 2 rRNA genes (rrnS and rrnL), 13 tRNA genes are also encoded on the heavy strand. However, the light strand (L-strand) was only predicted to contain eight tRNA genes.
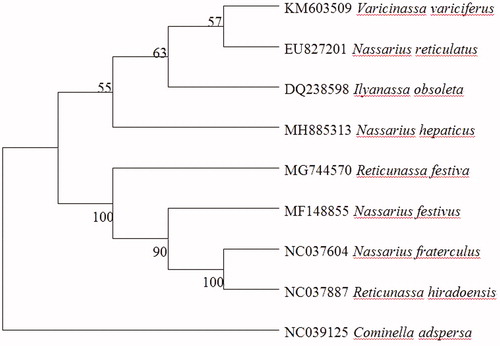
Phylogenetic analysis included mt genome of N. hepaticus and that of other eight species with high similarity. Maximum-likelihood (ML) analysis exhibited that N. hepaticus clustered with the Ilyanassa obsoleta (DQ238598), N. reticulates (EU827201), and Varicinassa variciferus (KM603509), highly supported by a bootstrap value of 55, Cominella adspersa (NC039125) was used as outgroup (). The evolutionary relationships of these analysed species are significant. The newly determined mitochondrial genome will help to understand the evolution of ratite.
Figure 1. Phylogenetic relationships (determined using the method of maximum likelihood) among the members of Nassarius hepaticus (Order: Mollusca) based on the Complete mitochondrial genome. The numbers beside the nodes are percentages of 1000 bootstrap values. The Cominella adspersa (NC039125) species was used as an outgroup. Alphanumeric terms indicate the GenBank accession numbers.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Bargues MD, Mas-Coma S. 1997. Phylogenetic analysis of Lymnaeid snails based on 18S rDNA sequences. Mol Biol Evol. 14:569–577.
- Cernohorsky WO. 1972. A taxonomic evaluation of recent and fossil non-mitrid species proposed in the family mitridae (mollusca: gastropoda). Rec Auck Inst Mus. 9:205–229.
- Li H, Lin D, Fang H, Zhu A, Gao Y. 2010. Species identification and phylogenetic analysis of genus Nassarius (Nassariidae) based on mitochondrial genes. Chin J Oceanol Limnol. 28:565–572.
- Passamaneck YJ, Schander C, Halanych KM. 2004. Investigation of molluscan phylogeny using large-subunit and small-subunit nuclear rRNA sequences. Mol Phylogenet Evol. 32:25–38.
- Wang W, Cai LZ, Liu WM. 2007. Morphological classification of nassariids in Fujian coast. J Xiamen Univ.6(A01):171–175. (Chinese paper)
