Abstract
The Shiba shrimp Metapenaeus joyneri is a commercially important species in China, Korea, and Japan. Thus far, genetic information, which is essential for the sustainable management of the resource, is quite limited for this species. In this study, the complete mitochondrial genome of M. joyneri was originally determined. Its mitogenome is 16,008 bp in length and contains 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and one control region (CR). The overall base composition is 35.5, 32.7, 19.4, and 12.4% for A, T, C, and G, respectively. All PCGs are initiated by ATN codons. Four of the 13 PCGs harbor the incomplete termination codon T. Phylogenetic analysis among the available malacostracans species supports the current morphology-based hypothesis that M. joyneri grouped with Metapenaeus species. These data provide fundamental information for further conservation genetic and phylogenetic studies of M. joyneri.
The shrimp Metapenaeus joyneri (Miers, 1880) is naturally distributed in the Indo-West Pacific region from Korea and Japan to southern China (Cha et al. Citation1995). Although this species contributes to commercial shrimp fisheries in these three countries, limited genetic information is currently available for M. joyneri, which hindered the establishment of effective management and conservation strategies. In this study, we characterized the complete mitochondrial genome of M. joyneri for the first time and explored the phylogenetic relationship among Malacostraca species. The results would be useful reference for further studies on resource conservation and management of this species.
The specimen was collected from Yellow Sea of China (33.011932°N, 122.015328°E), and stored at -80 °C prior to DNA extraction. DNA was isolated with phenol-chloroform method as described in Santos (Citation2006). All the specimen and DNA samples were deposited in the sample room of Division of Genetic Resources and Breeding, Yellow Sea Fisheries Research Institute. Eighteen pairs of primers were synthesized to amplify the whole mitogenome of M. joyneri. The transfer RNA (tRNA) genes in M. joyneri mitochondrial genome were recognized using MOTIS (Bernt et al. Citation2013). The gene map of M. joyneri mitogenome was generated using OGDraw v1.2 (Lohse et al. Citation2013). Phylogenetic analysis was carried out using Maximum Likelihood method with MEGA7.0 software (Kumar et al. Citation2016). The complete mitochondrial genome of M. joyneri was deposited in GenBank under Accession No. MH939247.
The complete genome sequence is 16,008 bp in length and contains 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes and one control region (CR). A total of 23 genes (62.1%) are encoded on the heavy strand while the remaining 14 genes (37.9%) are located on the light strand. The overall A + T content of the M. joyneri mitogenome is 66.2%, which is similar with that in other decapods (Karagozlu et al. Citation2018; Zhong et al. Citation2018). Twelve of the 13 protein-coding genes in the M. joyneri mitochondrial genome utilized the initiation codon ATG or ATT, which is common in decapod species (Meng et al. Citation2016; Kim et al. Citation2018). Among the protein-coding genes, nine (atp6, atp8, coxI, coxII, coxIII, cob, nad2, nad 3 and nad6) are encoded on the heavy strand while the remaining four (nad1, nad4L and nad4, nad5) are located on the light strand. The majority (9 of 13; 69.2%) of the protein-coding genes in the M. joyneri mitochondrial genome possess TAA or TAG as their termination codons, and the remaining 4 genes (coxI, coxII, coxIII, and nad3) terminate with T.
The mitochondrial rRNA genes of M. joyneri are located on the light strand, with 16S rRNA (1428 bp) flanked by tRNAIle (ATC) and tRNALeu (CAT) in the 5′ and 3′ directions, respectively, and 12S rRNA (863 bp) between tRNAVal (CTA) and CR. The location and orientation of these genes is relatively universal across the mitochondrial genomes in malacostracan species (Miller and Austin Citation2006; Ivey and Santos Citation2007). Fourteen of the 22 tRNA genes in the M. joyneri mitochondrial genome are located in five clusters, with each containing two or more tRNAs in a tandem arrangement and the remaining 8 tRNA genes are scattered in the mitogenome. The tRNA genes range from 65 to 72 bp in length. Fourteen of them are encoded on the heavy strand and the rest on the light strand.
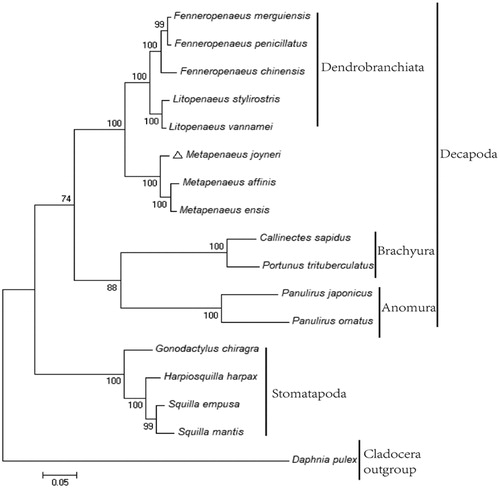
The phylogenetic position of M. joyneri within Decapoda was investigated with the concatenated alignments of amino acid in 17 decapod species using Maximum Likelihood method (). The result indicated that M. joyneri is clustered with other Metapenaeus species, which is in accordance with previous morphological analyses. The mitogenome resource obtained in the present study will form a valuable asset for improving the conservation of M. joyneri via further investigations on population genetics and provide useful reference for phylogenetic analyses of Decapoda.
Acknowledgements
We would like to thank Dr. Qiang Wu in Division of Fishery Resources and Ecosystem, Yellow Sea Fisheries Research Institute, for collecting the sample and identifying its species.
Disclosure statement
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. Mitos: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Cha HK, Choi JH, Oh CW. 1995. Reproductive biology and growth of the Shiba shrimp, Metapenaeus joyneri (Decapoda: Penaeidae), on the Western Coast of Korea. J Crustacean Biol. 24:93–100.
- Ivey JL, Santos SR. 2007. The complete mitochondrial genome of the Hawaiian anchialine shrimp Halocaridina rubra Holthuis, 1963 (Crustacea: Decapoda: Atyidae). Gene. 394:35–44.
- Karagozlu MZ, Dinh TD, Nguyen VQ, Kim CB. 2018. Analysis of complete mitochondrial genome of Etisus anaglyptus (Arthropoda, Decapoda, Xanthidae) with phylogenetic consideration. Mitochondr DNA B. 3:278–279.
- Kim NK, Andriyono S, Kim AR, Lee CI, Kim HW. 2018. Characterization of complete mitochondrial genome of two-spot swimming crab Charybdis bimaculata (Miers, 1886. ). Mitochondr DNA B. 3:902–903.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. Organellar Genome DRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Meng XL, Jia FL, Zhang XH, Liu P, Li J. 2016. Complete sequence and characterization of mitochondrial genome in the swimming crab Portunus sanguinolentus (Herbst, 1783) (Decapoda, Brachyura, Portunidae). Mitochondr DNA A. 27:3052–3053.
- Miller AD, Austin CM. 2006. The complete mitochondrial genome of the mantid shrimp Harpiosquilla harpax, and a phylogenetic investigation of the Decapoda using mitochondrial sequences. Mol Phylogenet Evol. 38:565–574.
- Santos SR. 2006. Patterns of genetic connectivity among anchialine habitats: a case study of the endemic Hawaiian shrimp Halocaridina rubra on the Island of Hawaii. Mol Ecol. 15:2699–2718.
- Zhong SP, Zhao Y, Wang XF, Song ZF, Zhang Q, Chen XL. 2018. The complete mitochondrial genome of the cryptic species (Form II) in kuruma shrimp Marsupenaeus japonicus (Decapoda: Penaeidae). Mitochondr DNA B. 3:184–186.