Abstract
We sequenced the complete mitogenome of Marenzelleria neglecta, a non-indigenous polychaete of the Baltic Sea originating from North America. This is the first complete mitogenome made available for the family Spionidae. The genome is 15,339 bp with a gene order identical to the generic order of the Pleistoannelida. The multigene maximum likelihood phylogeny, based on all protein-coding genes, weakly resolved the position of M. neglecta, as it tended to asssociate it with species representing the basal lineage of Annelids. This result underlines the need for more mitogenomes of Spionidae in order to improve the accuracy of their phylogeny.
Marenzelleria neglecta is a Polychaeta from the family Spionidae (Sikorski and Bick Citation2004). Although it originates from the Atlantic Coast of North America, Sikorski and Bick (Citation2004) used a specimen of the Southern Baltic Sea as holotype of M. neglecta during their revision of the genus. Marenzelleria spp. were identified as invasive species of brackish waters of the Baltic Sea in the mid 1980s. Due to their tolerance to a broad range of salinities (Bochert Citation1997; Sikorski and Bick Citation2004), they have become an important component of the local benthic macrofauna (Bick and Burckhardt Citation1989; Bastrop et al. Citation1997; Sikorski and Bick Citation2004). In the present study, we isolated total DNA from a single individual of M. neglecta that was collected in August 2018 from the Pomeranian Bay (53°55´37˝N 14°17´01˝E). Library construction and DNA sequencing on the BGISEQ-500 platform were carried out at the Beijing Genomic Institute (Shenzhen, China). The rest of the DNA sample is being kept in the University of Szczecin. A total of 50-millions paired-end reads of 100 bp were obtained and assembled using SPAdes 3.12.0 with a k-mer of 85. The 15,339-bp mitogenome (GenBank: MK120303) was recovered and annotated using Mitos (Bernt et al. Citation2013). It appears to be the first complete mitogenome made available for the family Spionidae. The M. neglecta mitogenome contains a total of 37 genes encoding 13 proteins, 2 rRNAs, and 22 tRNAs. The order of the protein-coding genes is identical to the generic order defined for the Pleistoannelida (Purschke Citation2015; Weigert et al. Citation2016). In the segment between atp8 and cox3, there is a tRNA gene cluster separated in two by a 614 bp intergenic spacer that may correspond to the control region.
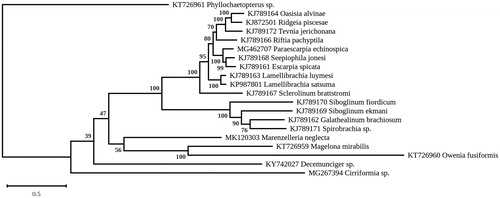
A RAxML (Stamatakis Citation2014) phylogenomic tree was inferred from the 13 protein-coding genes (atp6, atp8, cox1, cox2, cox3, cob, ND1, ND2, ND3, ND4, ND4L, ND5, ND6) of M. neglecta, other members of the infraclass Canalipalpata, a representative of the Magelonidae (Magelona mirabilis) and a member of the Chaetopteridae (Phyllochaetopterus sp.) as outgroup, with 1000 bootstrap replications and the GTR + I + G substitution model (). M. neglecta associated weakly with M. mirabilis and Owenia fusiformis, forming a clade that is sister to a strongly supported cluster mostly constituted of Sabellida. The placement of M. neglecta is unexpected because M. mirabilis and O. fusiformis are not part of the Pleistoannelids but rather represent a basal lineage of annelids (Purschke Citation2015; Weigert et al. Citation2016) and also because gene order in their mitogenomes differs from that of M. neglecta. Additional mitogenome sequences of Spionidae are clearly needed to resolve unambiguously the phylogenetic position of M. neglecta and in this regard, sampling of Marenzelleria viridis and Marenzelleria arctia from the Baltic Sea may prove useful.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Bastrop R, Röhner M, Sturmbauer C, Jürss K. 1997. Where did Marenzelleria spp. (Polychaeta: Spionidae) in Europe come from? Aquat Ecol. 31:119–136.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenetics Evol. 69:313–319.
- Bick A, Burckhardt R. 1989. Erstnachweis von Marenzelleria viridis (Polychaeta, Spionidae) für den Ostseeraum, mit einem Bestimmungsschlüssel der Spioniden der Ostsee. Mitt Zool Mus Berl. 2:237–247.
- Bochert R. 1997. Marenzelleria viridis (Polychaeta: Spionidae): a review of its reproduction. Aquat Ecol. 31:163–175.
- Purschke G. 2015 Annelida: basal groups and Pleistoannelida. In: Schmidt-Rhaesa A, Harzsch S, Purschke G. Structure and evolution of invertebrate nervous systems. Oxford, UK: Oxford University Press; p. 254–312
- Sikorski AV, Bick A. 2004. Revision of Marenzelleria Mesnil, 1896 (Spionidae, Polychaeta). Sarsia. 89:253–275.
- Stamatakis A. 2014. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Weigert A, Golombek A, Gerth M, Schwarz F, Struck TH, Bleidorn C. 2016. Evolution of mitochondrial gene order in Annelida. Mol Phylogenetics Evol. 94:196–206.