Abstract
Aspergillus parasiticus is a notorious filamentous fungus, which can produce aflatoxin B and G. Here, we reported the complete mitochondrial genome sequence of Aspergillus parasiticus isolated from air in South Korea. Its mitochondrial genome was successfully assembled from raw reads sequenced using MiSeq by Velvet and GapCloser. Total length of the mitochondrial genome is 29,141 bp and encoded 45 genes (17 protein-coding genes, 2 rRNAs, and 26 tRNAs). Nucleotide sequence of coding region takes over 25.4% and overall GC content is 26.2%. Phylogenetic tree presents that A. parasiticus is clustered with Aspergillus oryzae, which is same section Flavi in Aspergillus genus. It will be a useful molecular resource to understand section Flavi in Aspergillus genus.
Aspergillus parasiticus is a notorious filamentous fungus, which can produce aflatoxin B and G, which are poisonous carcinogens. The species is included in Secton Flavi, Aspergillus (Frisvad et al. Citation2019). Aspergillus sojae may be a domesticated species of A. parasiticus because A. sojae has no ability to produce mycotoxins so that it is used for making soybean sauce in Korea (Hong et al. Citation2015). Aspergillus flavus is another neighbour species, which can produce aflatoxin B (Gourama and Bullerman Citation1995). In the same section, Aspergillus oryzae and Aspergillus tamarii are domesticated and utilized in Japan (Ito et al. Citation1998). To understand genetic diversity and phylogenetic relationship of A. parasiticus in Section Flavi, we completed its mitochondrial genome.
A strain of A. parasiticus was collected from air in South Korea and its DNA was extracted by using a DNeasy Plant Mini kit (QIAGEN, Hilden, Germany). Raw data generated by MISeq were subject to de novo assembly done by Velvet 1.2.10 (Zerbino and Birney Citation2008) and gap filling with SOAPGapCloser 1.12 (Zhao et al. Citation2011) to get complete mitochondrial genome after confirming each bases using BWA 0.7.10 and SAM tools 1.9 (Li et al. Citation2009; Li Citation2013). Geneious R11 11.0.5 (Biomatters Ltd, Auckland, New Zealand) was used to annotate its chloroplast genome by comparing with that of Aspergillus luchuensis (MK061298; doi: 10.1080/23802359.2018.1547160). Voucher sample was deposited into KACC with accession KACC-46475.
The length of A. parasiticus mitochondrial genome (Genbank accession is MK124769) is 29,141 bp, which is shortest length among nine available Aspergillus mitochondrial genomes (Futagami et al. Citation2011; Joardar et al. Citation2012; Xu et al. Citation2018) (doi: 10.1080/23802359.2018.1547160). A. parasiticus COX1 gene, which contains several introns on fungal mitochondrial genomes (Joardar et al. Citation2012) has one intron sequence. In the intron, there is only one gene in A. parasiticus; while there are two genes in A. fumigatus (NC_017016; Joardar et al. Citation2012). Gene order of two mitochondrial genomes are same except that mitochondrial genome of A. fumigatus has additional two hypothetical ORFs (Joardar et al. Citation2012). A. parasiticus mitochondrial genome encoded 45 genes consisting of 17 protein-coding genes, 2 rRNAs, and 26 tRNAs. Nucleotide sequence of coding region takes over 25.4% and overall GC content is 26.2%, which is similar to mitochondrial genomes of Aspergillus genus.
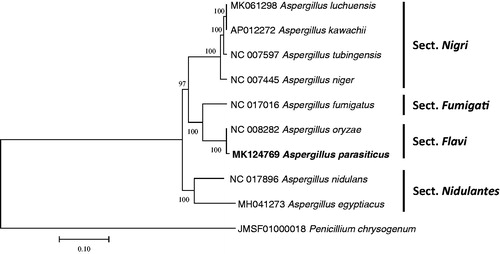
Sequence alignment of nine Aspergillus mitochondrial genomes and one Penicillium mitochondrial genome as an outgroup was conducted by MAFFT 7.388 (Katoh and Standley Citation2013). The neighbor joining was constructed using MEGA X with 10,000 bootstrap replicates (Kumar et al. Citation2018). Based on the phylogenetic tree, A. parasiticus is clustered with Aspergillus oryzae, which is same section Flavi in Aspergillus (Frisvad et al. Citation2019). This mitochondrial genome will be a useful molecular resource to understand section Flavi in Aspergillus genus, an important section for fermentation industry as well as fungal toxin.
Disclosure statement
The authors declare that they have no competing interests.
Figure 1. Neighbor joining phylogenetic tree (bootstrap repeat is 10,000) of nine Aspergillus mitochondrial genomes and one Penicillium mitochondrial genome: Aspergillus parasiticus (MK124769 in this study), Aspergillus luchuensis (MK061298), Aspergillus kawachii (AP012272), Aspergillus egyptiacus (MH041273), Aspergillus tubingensis (NC_007597), Aspergillus nidulans (NC_017896), Aspergillus niger (NC_007445), Aspergillus oryzae (NC_008282), Aspergillus fumigatus (NC_017016), and Penicillium chrysogenum (JMSF01000018). The numbers above branches indicate bootstrap support values of neighbor joining phylogenetic tree.
Additional information
Funding
References
- Frisvad JC, Hubka V, Ezekiel CN, Hong S-B, Nováková A, Chen AJ, Arzanlou M, Larsen TO, Sklenář F, Mahakarnchanakul W, et al. 2019. Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Stud Mycol. 93:1–63.
- Futagami T, Mori K, Yamashita A, Wada S, Kajiwara Y, Takashita H, Omori T, Takegawa K, Tashiro K, Kuhara S, Goto M. 2011. Genome sequence of the white koji mold Aspergillus kawachii IFO 4308, used for brewing the Japanese distilled spirit shochu. Eukaryot Cell. 10:1586–1587.
- Gourama H, Bullerman LB. 1995. Aspergillus flavus and Aspergillus parasiticus: Aflatoxigenic fungi of concern in foods and feeds: a review. J Food Prot. 58:1395–1404.
- Hong S-B, Kim D-H, Samson RA. 2015. Aspergillus associated with Meju, a fermented soybean starting material for traditional soy sauce and soybean paste in Korea. Mycobiology. 43:218–224.
- Ito Y, Peterson SW, Goto T. 1998. Properties of Aspergillus tamarii, A. caelatus and related species from acidic tea field soils in Japan. Mycopathologia. 144:169–175.
- Joardar V, Abrams NF, Hostetler J, Paukstelis PJ, Pakala S, Pakala SB, Zafar N, Abolude OO, Payne G, Andrianopoulos A, et al. 2012. Sequencing of mitochondrial genomes of nine Aspergillus and Penicillium species identifies mobile introns and accessory genes as main sources of genome size variability. BMC Genom. 13:698.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Preprint arXiv. 13033997:
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Xu Z, Wu L, Liu S, Chen Y, Zhao Y, Yang G. 2018. Structure characteristics of Aspergillus egyptiacus mitochondrial genome, an important fungus during the fermentation of dark tea. Mitochondrial DNA B. 3:1135–1136.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genom Res. 18:821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2.
