Abstract
Nomenclature for the nameless is one of the successful events after the invention of the DNA barcoding technique in biodiversity research. The ornamental fish species are frequently hunted from the east and northeast India and trafficked with various elusive names and high demanding values. Such illegitimate trading stimulated the severe threats on the native freshwater ecosystems and their indigenous biodiversity. Both traditional taxonomy and DNA barcoding technique successfully identified 11 ornamental fish species from a small riverine system, Murti river; linked up with three protected areas in the northern part of West Bengal. To test the efficacy of DNA barcode data for species identification, the generated sequences were subjected to similarity search results, and Neighbour-Joining tree clustering and genetic divergences. The mean genetic divergence was 21.3% and the interspecific genetic distance was ranging from 17.8% to 28.7% in the studied dataset. The detected high intraspecific genetic divergence in Opsarius barna (14.1%) and Channa gachua (6.4%) in the present dataset suggested further genetic investigation from their known distributions.
1. Introduction
Ornamental fishkeeping in an aquarium is a popular avocation and thus fetch high commercial value across the globe. In 2017, a total of 0.98 million USD incomes were made by freshwater ornamental fishes from India, which is around 0.47% as compared with the global export (MPEDA Citation2018). The eastern and northeastern region of India harbours an innumerable number of ornamental fishes from freshwater riverine and their tributaries (Bhattacharya and Choudhury Citation2004). In this region, 50% of extant fish species have potential economic value due to food production, tourism, and aquarium fish trade (Ponniah and Sarkar Citation2000; Das and Biswas Citation2008; Kalita and Deka Citation2013). Because of the commercial wealth and lack of breeding, many threatened and endemic ornamental fish species are being illegitimately traded by exporters from the natural habitats, and thus poses threat to the indigenous biodiversity. Furthermore, owing to the dearth of traditional taxonomic practices, the accurate identification of ornamental fishes is often challenging. Besides, the trade name of ornamental fishes always misleads to identify the species as various trade names were often used for a single species or vice versa (Dhar and Ghosh Citation2015). Nevertheless, the regional specific inventory of ornamental fishes helps to discover the living biological resources and predict the equity sharing through commercialized trade. Therefore, the intervention of molecular tools is essential to identify the ornamental fish species and monitoring their trade.
DNA barcoding is evidenced as an effective and additional tool in taxonomic research (Hebert et al. Citation2003; Hajibabaei et al. Citation2007). The technique has been successful to discover many new species (Mohapatra et al. Citation2013), resolve the taxonomic uncertainty (Laskar et al. Citation2013, Citation2018b), monitor the ornamental trade (Steinke et al. Citation2009; Collins et al. Citation2012), and biodiversity assessment (Ward et al. Citation2009; Laskar et al. Citation2018a) of fishes throughout the world, including India. Further, the molecular technique has proven to be useful to detect many non-native species (Kundu et al. Citation2016) and identify the on-going pet-trade of threatened taxa (Kundu et al. 2018a). In this study, we surveyed a small riverine system adjacent with three wildlife protected areas in West Bengal state of eastern India and applied the DNA barcoding tool for identifying the ornamental fish species from this region. Further, we tested the efficacy of the DNA barcode region to discriminate the studied species through genetic distance and Neighbour-Joining tree.
2. Materials and methods
2.1. Taxon sampling
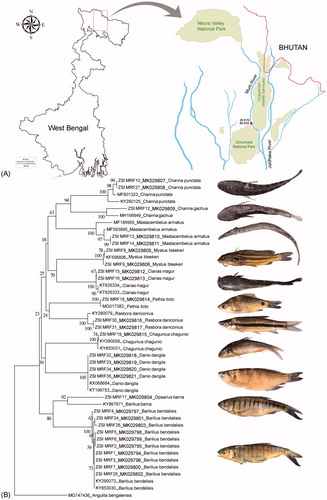
Total 11 ornamental fish species (Channa punctata = 2, Channa gachua = 1, Mastacembelus armatus = 2, Mystus bleekeri = 2, Clarias magur = 2, Pethia ticto = 1, Rasbora daniconius = 2, Chagunius chagunio = 1, Danio dangila = 4, Opsarius barna = 1, Barilius bendalisis = 10) were collected from the Murti River (26.81 N 88.83 E), in the northern part of West Bengal, India. The riverine system originates from the Neora Valley National Park (NP) and flows in-between Gorumara NP and Chapramari Wildlife Sanctuary (). The fish samples with desired size were collected and stored in 70% molecular grade alcohol without agitating the ecology and the biotic wealth of the riverine system. The studied species were identified through external morphological characters and meristic counts described previously (Vishwanath et al. Citation2014). The identified specimens were preserved with proper voucher numbers in National Zoological Collections (NZCs), Freshwater Fish Section, ZSI, FPS Building, Zoological Survey of India (ZSI), Kolkata. A small amount of muscle tissue from each specimen was collected aseptically for downstream molecular experiments. The excess amount of muscle tissues and genomic DNA were stored at the Centre for DNA Taxonomy Laboratory, Molecular Systematics Division, ZSI, Kolkata for future reference.
Figure 1. (A) Map with red dot showing the collection locality of ornamental fish species from Murti River. (B) Neighbour-joining (NJ) tree of the studied ornamental fish species with bootstrap supports. The database sequences of Anguilla bengalensis used as an out-group in the Neighbour-Joining tree. The photographs of the ornamental fish species were captured by second author and superimposed beside the species-specific clades in the Neighbour-Joining tree.
2.2. Genomic DNA extraction, amplification of mtCOI, and sequencing
Total genomic DNA was extracted from muscle tissue in 500 ml of TES buffer containing 50 mM Tris-HCl (pH 8.0), 25 mM EDTA (pH 8.0), 150 mM NaCl, and proteinase K (200 mg/ml) through a standard Phenol: Chloroform: Isoamyl Alcohol method (Sambrook and Russell Citation2001). The extracted DNA was checked on 1% agarose gel electrophoresis containing with Ethidium bromide. The published primer pair: FishF1: 5′-TCAACCAACCACAAAGACATTGGCAC-3′ and FishR1: 5′-TAGACTTCTGGGTGGCCAAAGAATCA-3′ was used to amplify the targeted partial mitochondrial Cytochrome C Oxidase Subunit I (mtCOI) gene fragment (Ward et al. Citation2005). The 30 ml reaction mixture contained 10 pmol of each primer, 100 ng of DNA template, 1× PCR buffer, 1.5 mM MgCl2, each dNTP at 0.25 mM, and 0.25 U of high-fidelity Taq DNA polymerase (Applied Biosystems Inc., Foster City, CA). The thermal profile for the PCR was set for an initial denaturation at 94 °C for two minutes, followed by 30 cycles of 94 °C for 45 seconds, 50 °C for 45 seconds, 72 °C for one minute and stored at 4 °C. PCR amplification was performed using a VeritiVR Thermal Cycler (Applied Biosystems Inc., Foster City, CA). The PCR product was purified using a QIAquickVR Gel Extraction Kit (QIAGEN Inc., Germantown, MD). Approximately, 10 ng of purified PCR products, both forward and reverse PCR primers were used for cycle sequencing, by using BigDyeVR Terminator ver. 3.1 Cycle Sequencing Kit (Applied Biosystems, Inc., Foster City, CA). Subsequently, the products from the cycle sequencing were cleaned using BigDye X-terminator Kit (Applied Biosystems Inc., Foster City, CA) and sequenced bi-directionally on a 48-capillary 3730 DNA Analyzer at the ZSI in-house sequencing facility.
2.3. Sequence quality check, dataset preparation and analysis
To obtain the consensus sequences of each specimen, both forward and reverse chromatograms were checked and trimmed at both the ends to discard the ambiguous bases and noisy part. The sequences were annotated carefully based on alignment of forward sequences and reverse complementary of reverse sequences of each sample. Finally, each of the sequences was compared in the GenBank database through nucleotide BLASTn (Basic Local Alignment Search Tool) search (https://blast.ncbi.nlm.nih.gov) and ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) to examine the accurate amino acid codes without any stop codon or indel (insertion/deletions). The similarity search of the generated sequences was performed through BLASTn and BOLD-IDs search in both GenBank and BOLD database. To make a combined dataset, the generated sequences and 17 published database sequences of same or related species were aligned through ClustalX software (Thompson et al. Citation1997). Further, to avoid the incongruous outcomes in genetic distance and Neighbour-Joining tree, the dataset was made to equal length and the database sequence of an eel fish species, Anguilla bengalensis was used as out-group. The evolutionary genetic divergence within and between the species was calculated in MEGAX (Kumar et al. Citation2018). To check the reciprocal monophyly among the studied species, Neighbor-Joining (NJ) tree was measured MEGAX.
3. Results and discussion
Due to the lacking of contemporary taxonomic intervention, the effective monitoring of the ornamental fish trade is not taking place in India and other countries (Murray et al. Citation2012). The aquarium fishes are commonly marketed by their trade names (Snakeheads and Barbs, etc.) instead of their ICZN zoological nomenclature and thus violated the employed regulations for many IUCN categorized threatened species (Raghavan et al. Citation2013). The present ichthyological survey collected total 11 ornamental fish species from the short stretch of freshwater eco-system in Murti River. The collected ornamental fishes were barcoded and submitted in both GenBank and BOLD data system under the project ‘DNA barcoding of Indian ornamental fishes’. The global database with DNA barcode similarity search engine (BLASTn and BOLD-IDs) offers a quick access in species identification based upon the nearest match of the query sequences. Most of the morphologically identified ornamental fish species revealed 99% to 100% similarity with the representative database sequences of the same species. The B. barna is synonymous with O. barna, thus the result is considered as straightforward. Further, the resulted Neighbour-Joining tree shows distinct clustering of all the studied species in the present dataset with their representative DNA barcode sequences (). The genetic distances discussed here revealed from the present dataset and might be altered for others. The mean genetic divergence of the studied dataset was 21.3%. The intraspecific genetic distances range from 0% (Danio dangila) to 14.1% (O. barna) in the present dataset. The high intraspecific genetic divergence in O. barna (14.1%) and Channa gachua (6.4%) were observed in the studied dataset as compared with the database sequences (B. barna KY867671 and C. gachua MH156949), generated from the nearest northeastern region of India. Thus, the present study suggested further genetic investigation from their type locality and other broad geographical areas. Moreover, a detailed survey of freshwater fish species in the studied locality required to determine the presence of possible species complexes and cryptic diversity. The interspecific genetic distance in the studied dataset ranged from 17.8% to 28.7%. The O. barna, previously known as B. barna shows 18.4% genetic divergence with Barilius bendalisis. However, the two congeners of Channa, C. punctata and C. gachua show 20% genetic divergence in the studied dataset. The aimed study successfully identified the ornamental fishes that are reported to be commercialized and frequently traded in eastern and northeastern part of India.
The study successfully identified the ornamental fish species from an important river in east India by using both traditional taxonomy and contemporary molecular approaches. Importantly, this river flows through three protected areas, the popular conservation sites for Indian Rhinoceros, Royal Bengal Tiger, Gaur, Asian elephant, Red Panda, Bear, and Deer. The river originates from the Neora Valley NP and move through the medium gradient hills, and finally join with the larger stream of River Jaldhaka. The study aimed to identify the ornamental fish diversity in Murti River is an important genetic study, because, in its short stretch, before adjoining into the River Jaldhaka, sustains a less agitated riparian conditions. Indeed, being connected with the NP, the river stretch is often protected by the forest departments to restrict illegal fishing and to preserve pristine natural habitats. Notably, this is an example of prior importance to generate DNA barcode data of the extant biota from the protected areas and conservation sites.
The DNA barcoding tool has proven to be effective for identification of many animal taxa from different reliable source of biological samples (blood, muscle tissue, hairs, stool, and environmental samples etc.) (Kundu et al. Citation2018b). The molecular technique can also identify the species from different life stages, amorphous samples, cooked meat, or from packaged food materials. Thus, the technique is useful in systematics research by resolving many taxonomic quests in one side, and also helps to detect many unlawful trades, adulteration, or fraud cases on another side. Hence, the present effort indicated that the molecular methods might play an important role in regulating the uncontrolled exploitation of ornamental fishes. The study further demanded to implement more strategic conservation action plan for sustaining the threatened or endemic ornamental fish species in their natural habitats.
Acknowledgements
The authors are thankful to the Director of Zoological Survey of India (ZSI), Ministry of Environment, Forests and Climate Change (MoEF&CC), Govt. of India for providing necessary facilities, constant support and encouragement throughout the study. We are thankful to the West Bengal Biodiversity Board for their support to L.K. and U.D. We are also acknowledge to the ‘NMHS large grant, Conservation of Threatened Vertebrate Fauna in Indian Himalayan Region through Long-Term Monitoring and Capacity Building’ to K.C. and S.K.; and ZSI, MoEF&CC Core Funding to V.K. and K.T. The funders had no role in study design, data collection and analysis or preparation of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bhattacharya BK, Choudhury M. 2004. Ornamental Fish Culture and Trade in Northeastern India. Kolkata: Central Inland Fisheries Research Institute (CIFRI).
- Collins RA, Armstrong KF, Meier R, Yi Y, Brown SD, Cruickshank RH, Keeling S, Johnston C. 2012. Barcoding and border biosecurity: identifying cyprinid fishes in the aquarium trade. PLoS One. 7:e28381.
- Das SP, Biswas JN. 2008. A Handbook of ornamental fishes of the Brahmaputra basin. Guwahati, India: Eastern Book House; 109 p.
- Dhar B, Ghosh SK. 2015. Genetic assessment of ornamental fish species from North East India. Gene. 555:382–392.
- Hajibabaei M, Singer GA, Hebert PD, Hickey DA. 2007. DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends Gen. 23:167–172.
- Hebert PD, Cywinska A, Ball SL, deWaard JR. 2003. Biological identifications through DNA barcodes. Proc Royal Soc B Biol Sci. 270:313–321.
- Kalita T, Deka K. 2013. Ornamental fish conservation in the flood plain wetlands of lower Brahmaputra Basin. Adv Appl Sci Res. 4:99–106.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Kundu S, Kumar V, Laskar BA, Chandra K, Tyagi K. 2016. Mitochondrial DNA effectively detects non-native Testudines: invisible wildlife trade in northeast India. Gene Rep. 4:10-15. DOI:10.1016/j.genrep.2016.02.002.
- Kundu S, Kumar V, Laskar BA, Tyagi K, Chandra K. 2018a. Pet and turtle: DNA barcoding identified twelve Geoemydid species in northeast India. Mitochondrial DNA Part B. 3:513–518.
- Kundu S, Kumar V, Tyagi K, Chandra K. 2018b. Environmental DNA (eDNA) testing for detection of freshwater turtles in a temple pond. Herpetol Notes. 11:369–371.
- Laskar BA, Bhattacharjee MJ, Dhar B, Mahadani P, Kundu S, Ghosh SK. 2013. The species dilemma of Northeast Indian Mahseer (Actinopterygii: Cyprinidae): DNA barcoding in clarifying the riddle. PLoS One. 8:e53704.
- Laskar BA, Kumar V, Kundu S, Darshan A, Tyagi K, Chandra K. 2018a. DNA barcoding of fishes from River Diphlu within Kaziranga National Park in northeast India. Mitochondrial DNA Part A. 18:1–9.
- Laskar BA, Kumar V, Kundu S, Tyagi K, Chandra K. 2018b. Taxonomic quest: validating two mahseer fishes (Actinopterygii: Cyprinidae) through molecular and morphological data from biodiversity hotspots in India. Hydrobiologia. 815:113–124.
- Mohapatra A, Ray D, Kumar V. 2013. A new fish species of the Genus Hapalogenys (Perciformes: Hapalogenyidae) from the Bay of Bengal, India. Zootaxa. 3718:367–377.
- MPEDA (Marine Products Export Development Authority). 2018. Data Source: UN Comtrade [Accessed 2018 Nov 11]. http://fishexchange.mpeda.gov.in.
- Murray JM, Watson GJ, Giangrande A, Licciano M, Bentley MG. 2012. Managing the marine aquarium trade: revealing the data gaps using ornamental polychaetes. PLoS One. 7:e29543.
- Ponniah AG, Sarkar UK. 2000. Fish biodiversity of North East India. NBFGR's NATP Special Publication No. 2. Nat Bureau of Fish Gen Res. 2:1-228.
- Raghavan R, Dahanukar N, Tlusty MF, Rhyne AL, Krishna Kumar K, Molur S, Rosser AM. 2013. Uncovering an obscure trade: threatened freshwater fishes and the aquarium pet markets. Biol Conserv. 164:158–169.
- Sambrook J, DW. Russell 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press.
- Steinke D, Zemlak TS, Hebert PD. 2009. Barcoding nemo: DNA-based identifications for the ornamental fish trade. PLoS One. 4:e6300.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882.
- Vishwanath W, Nebeshwar K, Lokeshwar Y, Shangningam BD, Rameshori Y. 2014. Freshwater fish taxonomy and a manual for identification of fishes of North-east India. Imphal, Manipur, India: Manipur University & National Bureau of Fish Genetic Resources.
- Ward RD, Hanner R, Hebert PD. 2009. The campaign to DNA barcode all fishes, FISH-BOL. J Fish Biol. 74:329–356.
- Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. 2005. DNA barcoding of Australia’s fish species. Philosophical Transactions of the Royal Society B. 360:1847–1857.
