Abstract
In order to supply genetic information of Acer truncatum Bunge, we reported the complete chloroplast genome sequence based on high-throughput sequencing data. The size of chloroplast genome is 156,262 bp length with overall GC content 37.9%. It exhibited typical quadripartite structure with 86,018 bp of LSC region, 18,072 bp of SSC region, and 26,086 bp of each IR region. A total of 137 genes were identified, including 89 protein-coding genes, 40 tRNA genes, and 8 rRNA genes. Phylogenetic analysis showed that A. truncatum was closely related to the congeneric A. miaotaiense.
Acer truncatum Bunge (Aceraceae), a maple native species to northern China, is a vulnerable tree species which has important ecological and medical value (Li et al. Citation2015). It has also been used as a Chinese folk medicine for the treatment of angina pectoris, coronary artery cirrhosis, and cerebrovascular diseases (Ma et al. Citation2005). The wild resources of A. truncatum declined significantly due to anthropogenic disturbance in recent years, thus, it has been listed as a nearly threatened species (Yin et al. Citation2018). It is urgently to adopt corresponding measures to protect wild germplasm. Here, we assembled and characterized the chloroplast (cp) genome of A. truncatum, which might provide genetic information relevant to the effective conservation and management of this species.
Fresh leaves of A. truncatum were collected from Mt. Heikuang in Yantai (N37°30′36.72ʺ, E121°24′35.99ʺ), Shandong Province, China. A voucher specimen was deposited in Northwest A&F University Herbarium. The genomic DNA was extracted using a DNeasy Plant Mini Kit (QIAGEN, Valencia, CA). The whole-genome sequencing was conducted with 150 bp pair-end reads on the Illumina Hiseq Platform (Illumina, San Diego, CA). The resulting contigs were linked based on overlapping regions after being aligned to the cp genome of the congeneric A. davidii (KU977442) (Jia et al. Citation2016) and visualized in Geneious R9 (Biomatters Ltd., Auckland, New Zealand). Annotation was conducted using the program Plann (Huang and Cronk Citation2015). A physical map of the cp genome was generated using OGDRAW (http://ogdraw.mpimp-golm.mpg.de/). Finally, the complete cp genome sequence was submitted to GenBank under the accession number of MG209700. To ascertain the phylogenetic position of A. truncatum within the genus Acer, its cp genome was multi-aligned with seven available cp genomes using MAFFT (Katoh and Standley Citation2013) and a neighbour joining (NJ) phylogenetic tree was constructed in MEGA6 (Tamura et al. Citation2013).
The cp genome of A. truncatum is a typical quadripartite structure with a length of 156,262 bp, which contained two inverted repeats (IR) of 26,086 bp separated by a large single-copy (LSC) and a small single-copy (SSC) of 86,018 bp and 18,072 bp, respectively. The cp genome contained 137 genes, comprising 89 protein-coding genes (PCGs), 40 tRNA genes, 8 rRNA genes. The genome contained 91 unique genes, 20 genes duplicated in the IRs and three species (trnG-UCC, trnM-CAU, and trnT-GGU) reside within the LSC region. Among the annotated genes, 15 of them contain one intron, which including 9 PCGs (ndhA, ndhB, petB, petD, atpF, rpl2, rpl16, rpoC1, and rps16) and 6 tRNAs (trnI-GAU, trnA-UGC, trnK-UUU, trnG-UCC, trnL-UAA, and trnV-UAC), and 3 genes (rps12, clpP, and ycf3) possess two introns. The overall GC content of A. truncatum cp genome was 37.9%.
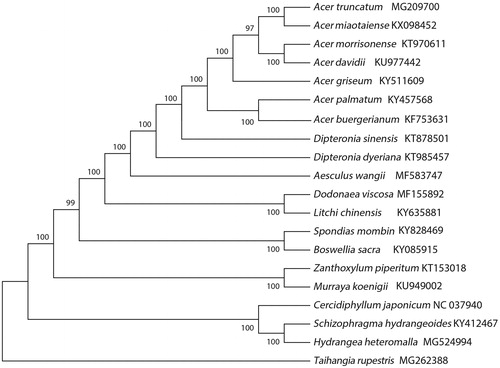
The phylogenetic analysis of 20 cp genomes in which Taihangia rupestris (MG262388) was selected as the outgroup displayed that A. truncatum was closely related to the A. miaotaiense (). The complete cp genome of A. truncatum will supply useful genetic information for population genetic survey, conduct phylogenetic analysis, evolutionary studies, and conservation strategies for this valuable tree species.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Jia Y, Yang J, He Y-L, He Y, Niu C, Gong L-L, Li Z-H. 2016. Characterization of the whole chloroplast genome sequence of Acer davidii Franch (Aceraceae). Conservation Genet Resour. 8:141–143.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Li L, Manning WJ, Tong L, Wang X. 2015. Chronic drought stress reduced but not protected Shantung maple (Acer truncatum Bunge) from adverse effects of ozone (O3) on growth and physiology in the suburb of Beijing, China. Environ Pollut. 201:34–41.
- Ma XF, Wu LH, Ito Y, Tian WX. 2005. Application of preparative high-speed counter-current chromatography for separation of methyl gallate from Acer truncatum Bunge. J Chromatogr A. 1076:212–215.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Yin DD, Li SS, Wu Q, Feng CY, Li B, Wang QY, Wang LS, Xu WZ. 2018. Advances in research of six woody oil crops in China. Chin Bull Bot. 53:110–125.