Abstract
Juniperus tibetica forms the highest known treeline in the Northern Hemisphere at 4900 m a.s.l., and it is categorized as ‘vulnerable’ in the IUCN Red List of Threatened Species. Here, Illumina pair-end reads of Juniperus tibetica was used to characterize its complete chloroplast (cp) genome sequences. The circular genome with 127,662 bp in length contains two duplicated genes (trnI-CAU and trnQ-UUG), and 119 single copy genes that including 82 protein-coding genes, 33 transfer RNA and four ribosomal RNA genes. There are no inverted repeat sequences in this genome. The GC content of this genome is 35.04%. The phylogenomic analysis strongly supported the monophyly of both Juniperus sect. Juniperus and J. sect. Sabina; within J. sect. Sabina, J. tibetica is sister to all other involved species.
Juniperus tibetica Kom. (Cupressaceae) is an endemic tree of the southern Qinghai-Tibet Plateau (Farjon Citation2005; Lars Opgenoorth Citation2009), which was categorized as ‘vulnerable’ in the Red List of Threatened Species (Farjon Citation2013). This species forms the highest known treeline in the Northern Hemisphere at 4900 m a.s.l. To facilitate the preservation of this species, it is necessary to survey its genetic background.
Chloroplast genome consists of two inverted repeat (IR) sequences which separate genome into large (LSC) and small single copy (SSC) regions (Jansen and Ruhlman Citation2012). However, some clades of gymnosperm such as conifer lost inverted repeats, which lead to gene loss in cp genomes (Wu et al. Citation2011; Li et al. Citation2016a,Citationb). In our study, we assembled and characterized the complete cp genome sequences of J. tibetica using Illumina pair-end sequencing reads.
The leaf samples were collected from Linzhou County (Tibet, China; coordinates: 30°18.58′N, 91°30.72′E), and total genomic DNA was extracted from leaf tissues with modified CTAB method (Doyle Citation1987). The whole-genome sequencing was conducted on the Illumina Hiseq Platform (Illumina, San Diego, CA) and 19.84M 150-bp raw paired reads were yielded. After removing the adapters, the complete cp genome was assembled via NOVOplasty (Dierckxsens et al. Citation2017) using the cp genome of J. monosperma (Guo et al. Citation2014) as a reference. The cp genome was annotated using Plann (Huang and Cronk Citation2015) and annotation was corrected using Geneious (Kearse et al. Citation2012). The genome map was generated by using the web server OGDRAW (https://chlorobox.mpimp-golm.mpg.de/) (Lohse et al. Citation2013). The complete cp genome sequences were deposited in NCBI GenBank under Accession Number MK135439.
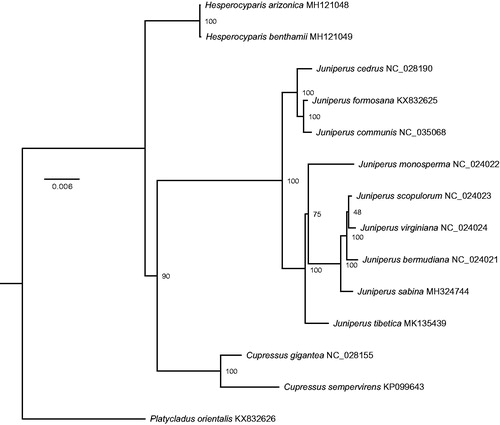
For phylogenetic analysis, cp genome sequences of eight Juniperus, two Cupressus, two Hesperocyparis species and Platycladus orientalis were downloaded from National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/), and 79 genes were extracted from their cp genome sequences. After concatenated and aligned by MAFFT (Katoh et al. Citation2002), the alignment was used to construct maximum likelihood (ML) tree via RAxML (Stamatakis Citation2014) with 200 bootstrap replicates.
The complete cp genome of J. tibetica is a double-stranded, circular DNA with 127,662 bp length that contains 119 single copy genes, including 82 protein-coding genes, 33 tRNA genes, and four rRNA genes. It also contains two genes (trnI-CAU and trnQ-UUG) occurred in double copies. Nine genes have a single intron, one gene harbor two introns. Further, this cp genome does not have canonical inverted repeat (IR) sequences, which is similar to another cp genome of conifers (Zhang et al. Citation2017). The overall GC content of J. tibetica complete cp genome is 35.04%.
The phylogenetic analysis using 14 chloroplast complete genomes of four genera showed that all involved Juniperus species clustered into two supported monophyletic groups, which correspond to Juniperus sect. Juniperus and J. sect. Sabina, respectively. Within the latter, J. tibetica is sister to the other five species ().
Figure 1. Phylogenetic relationships between the genus Juniperus and other three genera based on the maximum likelihood (ML) analysis of 14 complete chloroplast genome sequences from 9 Juniperus, 2 Cupressus, 2 Hesperocyparis, and 1 Platycladus. The supporting values based on 200 replicates are shown behind the nodes.
The complete chloroplast genome of this vulnerable conifer, J. tibetica, can provide information for the conservation for this species. It is also useful to the phylogenetic studies of conifers.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Farjon A. 2005. A monograph of Cupressaceae and Sciadopitys. London: Royal Botanic Garden, Kew.
- Farjon A. 2013. Juniperus tibetica. The IUCN Red List of Threatened Species 2013: e.T42256A2967451; [accessed on 05 Nov 2018]. https://www.iucnredlist.org/species/42256/2967451
- Guo WH, Grewe F, Clark AC, Fan WS, Duan ZL, Adams RP, Schwarzbach AE, Mower JP. 2014. Predominant and substoichiometric isomers of the plastid genome coexist within juniperus plants and have shifted multiple times during cupressophyte evolution. Genome Biol Evol. 6:580–590.
- Huang DI, Cronk QC. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Jansen RK, Ruhlman TA. 2012. Plastid genomes of seed plants. Dordrecht: Springer.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Lars Opgenoorth. 2009. Identification and characterization of microsatellite marker in the tetraploid Juniperus tibetica Kom. using next generation sequencing. Conserv Genet Res. 1:251–255.
- Li HE, Guo QQ, Zheng WL. 2016a. The complete chloroplast genome of Cupressus gigantea, an endemic conifer species to Qinghai-Tibetan Plateau. Mitochondrial DNA. 27:3743–3744.
- Li ZH, Qian ZQ, Liu ZL, Deng TT, Zu YM, Zhao P, Zhao GF. 2016b. The complete chloroplast genome of Armand pine Pinus armandii, an endemic conifer tree species to China. Mitochondrial DNA A DNA Mapp Seq Anal. 27:2635–2636.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wu CS, Lin CP, Hsu CY, Wang RJ, Chaw SM. 2011. Comparative chloroplast genomes of pinaceae: insights into the mechanism of diversified genomic organizations. Genome Biol Evol. 3:309–319.
- Zhang ZL, Ma LY, Yao H, Yang X, Luo JH, Gong X, Wei SY, Li QF, Wang W, Sun HB. 2017. The complete chloroplast genome of Cupressus chengiana. Conserv Genet Res. 9:1–3.
