Abstract
Cryptopone sauteri Wheeler, W.M., 1906, which is one of the common ant species in Ponerine, is found in forest floors of the Korean peninsula. We have determined the mitochondrial genome of C. sauteri. The circular mitogenome of C. sauteri is 15,367 bp including 13 protein-coding genes, two ribosomal RNA genes, 22 transfer RNAs, and a single large non-coding region of 448 bp. The base composition was AT-biased (82.0%). Gene order of C. sauteri is almost same to that of Cardiocondyla obscurior even though the two species are in different subfamilies. Phylogenetic tree agrees with the current phylogenetic placement of Cryptopone outside of all other ant mitochondrial genomes and in its own Poneroid clade. C. sauteri mitochondrial genome will be a useful resource for further analyses.
The Poneroid clade is a major ant taxon made up of 1,532 species belonging to six subfamilies (Bolton Citation2018). This clade has been studied with two major topics: i) the evolutionary history due to phylogenic importance (Borowiec et al. Citation2017) and ii) evolution of social structures due to their primitive society (Schmidt and Shattuck Citation2014). Despite of these interesting research topics, genomic resources are not enough: there are two whole genome sequences (Bonasio et al. Citation2010; Patalano et al. Citation2015), while there is no mitochondrial genome in this clade.
Cryptopone sauteri belonging to subfamily Ponerinae is commonly found in Korean peninsula. It has clear characteristics easily identified from other Korean Ponerine ants: its orange body color and setae on the outer surfaces of middle tibiae. Cryptopone sauteri is a hypogenic hunter species of the forest floor feeding on small invertebrates such as beetle and fly larvae (Group and Imai Citation2003). Its nests and founder queens are mostly found in rotting wood and leaf litter.
As a first step to understand genomic characteristics of Ponerine ants, we completed mitochondrial genome of C. sauteri isolated from Jeju Island, Republic of Korea. Genomic DNA was extracted from worker ants and sequenced by HiSeq4000 (Macrogen Inc, Seoul, Korea). Raw sequences were filtered by Trimmomatic 0.33 (Bolger et al. Citation2014) and de novo assembled by Velvet 1.2.10 (Zerbino and Birney Citation2008). Gaps were filled by SOAPGapCloser 1.12 (Zhao et al. Citation2011) and confirmed with BWA 0.7.17 and SAMtools 1.9 (Li et al. Citation2009; Li Citation2013). Geneious R11 11.1.5 (Biomatters Ltd, Auckland, New Zealand) was used to annotate its genome based on that of Cardiocondyla obscurior (KX951753). ARWEN (Laslett and Canbäck Citation2008) was used to annotate tRNAs. DNA sample of these ants was deposited in InfoBoss Cyber Herbarium (IN; Republic of Korea; J. Park, KFDS00047).
Cryptopone sauteri mitochondrial genome length (Genbank accession is MK138572) is 15,367 bp. The nucleotide composition is AT-biased (82.0%). The mitogenome contains 13 protein-coding genes (PCGs), two rRNAs, and 22 tRNAs. The tRNAs size ranges from 56 to 78 bp, are similar to other ants (circa 54–90 bp). Gene order of C. sauteri is almost identical to C. obscurior except the position of tRNA-Val, even though both species are in different subfamilies. The control region presumably corresponds to the single largest non-coding AT-rich region (448 bp, A+T 86.4%).
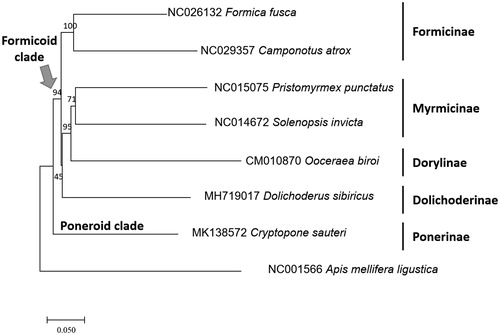
We inferred the phylogenetic relationship of seven ants including C. sauteri and one outgroup species using concatenated nucleotide sequences of all PCGs. Multiple sequence alignment was conducted by ClustalW (Thompson et al. Citation2003). Neighbour-joining tree was constructed using MEGA X with 10,000 bootstrap replicates (Kumar et al. Citation2018) presenting the phylogenetic position of C. sauteri based on mitochondrial genomes (). Results agree with current phylogenetic placement with Cryptopone placed sister to all Formicoid ants (). As the first species of which mitochondrial genomes are available in the subfamily Ponerinae, C. sauteri mitochondrial genome will be used for understanding molecular phylogenic relationship of Poneroid clade together with already sequenced mitochondrial genomes of ants in Formicoid clade including Dolichoderus sibiricus (Park et al., doi:10.1080/23802359.2018.1551091).
Figure 1. Phylogenetic tree of Cryptopone sauteri (This study; MK138572), as well as representative species of all available ant subfamilies: Ooceraea biori (CM010870), Dolichoderus sibiricus (MH719017), Camponotus atrox (NC_029357), Formica fusca (NC_026132), Solenopsis Invicta (NC_014672), Pristomyrmex punctatus (NC_015075), and a honey bee, Apis mellifera ligustica (NC_001566) as an outgroup. The numbers above branches indicate bootstrap support values.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Bolton B. 2018 An online catalog of the ants of the world. Available from http://antcat.org (accessed 2018 May 03).
- Bonasio R, Zhang G, Ye C, Mutti NS, Fang X, Qin N, Donahue G, Yang P, Li Q, Li C, et al. 2010. Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator. Science. 329:1068–1071.
- Borowiec ML, Rabeling C, Brady SG, Fisher BL, Schultz TR, Ward PS. 2017. Compositional heterogeneity and outgroup choice influence the internal phylogeny of the ants. bioRxiv.173393.https://www.biorxiv.org/content/early/2017/08/08/173393
- Group JAD, Imai HT. 2003. Ants of Japan. Japan: Gakken.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Patalano S, Vlasova A, Wyatt C, Ewels P, Camara F, Ferreira PG, Asher CL, Jurkowski TP, Segonds-Pichon A, Bachman M, et al. 2015. Molecular signatures of plastic phenotypes in two eusocial insect species with simple societies. Proc Natl Acad Sci. 112:13970–13975.
- Schmidt CA, Shattuck SO. 2014. The higher classification of the ant subfamily Ponerinae (Hymenoptera: Formicidae), with a review of ponerine ecology and behavior. Zootaxa. 3817:1–242.
- Thompson JD, Gibson TJ, Higgins DG. 2003. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics.2.3.1-2.3.22. https://currentprotocols.onlinelibrary.wiley.com/doi/pdf/10.1002/0471250953.bi0203s00
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 2011. 12:S2.
