Abstract
In the present study, the complete mitochondrial genome (mitogenome) sequence of Coilia brachygnathus collected from Huaihe river, was determined by long PCR and primer walking methods. Similar to other fishes, the mitogenome is a circular molecule 16,896 bp in length and contains 13 protein-coding genes (PCGs), 2 rRNA genes, 22 transfer RNA genes, and a control region (CR). In addition, the extended termination associated sequence domain (ETAS), the Central conserved domain (CSB-F, CSB-E, and CSB-D) and the Conserved sequence block (CSB1, CSB2, and CSB3) were identified in the control region. Phylogenetic analysis showed that C. brachygnathus first clustered together with C. nasus among Coilia fishes and formed a monophyly. The results will helpful for further understanding classification, phylogenetics, and biogeography of the Coilia fishes.
Coilia brachygnathus (Kreyenberg and Pappenheim 1908), belongs to Engraulidae, Clupeiformes, and is a small-to moderate-sized freshwater fish. It is widely distributed in the rivers with accessory lakes in the south central of China. Coilia brachygnathus is a disputed species, and some researchers believe that C. brachygnathus is an ecotype of Coilia nasus rather than a valid species because of similar morphological characteristics and DNA sequences between them (Cheng and Han Citation2004; Tang et al. Citation2007; Xu et al. Citation2009; Zhou et al. Citation2010). However, the complete mitogenome of C. brachygnathus from Huaihe River is still unknown up to now. The results of our present study will be helpful for further understanding of classification, phylogenetics, and biogeography of the Coilia fishes.
In this study, the complete mitogenome sequence of C. brachygnathus collected from Huaihe river (32°41′24ʺN, 116°42′36ʺE; Fengtai county, Anhui province, China, September 2018), was determined by long PCR and primer walking methods and was deposited in GenBank database with accession number MK134714. The specimen is stored at −70 °C in our laboratory (Anhui Provincial Key Laboratory of Aquaculture & Stock Enhancement). Similar to other fishes (Broughton et al. Citation2001; Li et al. Citation2016; Zhang and Xian Citation2016), the mitogenome is a circular molecule of 16,896 bp in length and contains 13 protein-coding genes (PCGs, 11,411 bp in all), 2 rRNA genes, 22 tRNA genes and a control region (CR). The encoding genes of mitogenome were located on H-strand except for ND6 and 8 tRNA (Gln, Ala, Asn, Cys, Tyr, Ser-UCN, Glu, and Pro) encoded on L-strand. 12S rRNA and 16S rRNA are 953 bp and 1688 bp in size, respectively. Twenty-two tRNA interspersed between the rRNAs and PCGs were identified, ranging in size from 66 bp (tRNACys) to 75 bp (tRNALeu). The origin of L-strand replication (OL) between the tRNAAsn and tRNACys could form a stable stem-loop secondary structure. The largest non-coding region (CR, 1252 bp in length) is located between the tRNAPro and tRNAPhe genes.
Compared with some studies on the sequence structure of CR (Tang et al. Citation2005; Zhu et al. Citation2008; Zhao et al. Citation2016), the extended termination associated sequence domain (ETAS), the Central conserved domain (CD) and the Conserved sequence block (CSB) were identified in CR of C. brachygnathus. Further, we derived the termination-associated sequence with the consensus sequence as TACATAT – ATGTATTATAT in the ETAS domain, 3 conserved sequences (CSB-F, CSB-E, and CSB-D) in CD domain and 3 conserved sequences (CSB-1, CSB-2, and CSB-3) in the CSB domain.
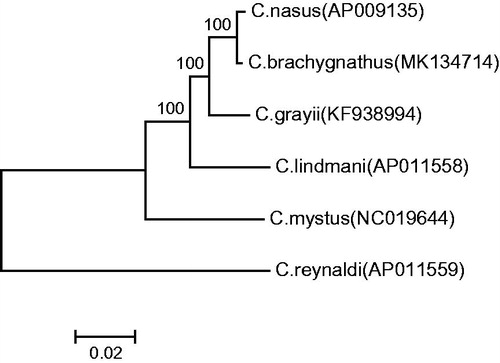
Phylogenetic analysis was performed by MEGA 6 (Tamura et al. Citation2013) based on the whole mitogenome sequences of 6 species in the genus Coilia. The neighbour-joining tree () showed that C. brachygnathus was first clustered together with C. nasus and formed a monophyly, and then they successively clustered together with other species: Coilia grayii, Coilia lindmani, Coilia mystus, Coilia reynaldi.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Broughton RE, Milam JE, Roe BA. 2001. The complete sequence of the zebrafish (Danio rerio) mitochondrial genome and evolutionary patterns in vertebrate mitochondrial DNA. Genome Res. 11:1958–1967.
- Cheng QQ, Han JD. 2004. Morphological variations and discriminant analysis of two populations of Coilia ectenes. J Lake Sci. 16:356–364.
- Li Y, Gao TX, Song N. 2016. The complete mitogenome of Pampus argenteus (Perciformes: Stromateidae). Mitochondrial DNA Part A. 27:684–685.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Tang WQ, Hu XL, Yang JQ. 2007. Species validities of Coilia brachygnathus and C. nasus taihuensis based on sequence variations of complete mtDNA control region. Biodivers Sci. 15:224–231.
- Tang QY, Liu HZ, Yang XP, Xiong BX. 2005. Studies on the structure of the mitochondrial DNA control region and phylogenetic relationships of the subfamily Botiinae. Acta Hydrobiologica Sinica. 29:645–653.
- Xu ZQ, Ge JC, Huang C, Dou HX, Pan JL, Xia AJ. 2009. Taxonomy of short jaw tapertail anchovy Coilia brachygnathus by jaw length and mitochondrial Cytochrome b gene analysis. J Dalian Fish Univ. 24:242–246.
- Zhang H, Xian WW. 2016. The complete mitochondrial genome of the larvae Japanese grenadier anchovy Coilia nasus (Clupeiformes, Engraulidae) from Yangtze estuary. Mitochondrial DNA Part A. 27:852–853.
- Zhao LL, Zhao YH, Zhang N, Gao TX, Zhang ZH. 2016. The complete mitogenome of Coilia nasus (Clupeiformes, Engraulidae) from Poyang Lake. Mitochondrial DNA Part A. 27:1608–1609.
- Zhou XD, Yang JQ, Tang WQ, Dong LIU. 2010. Species validities analyses of Chinese Coilia fishes based on mtRNA COI Barcoding. Acta Zootaxonomica Sinica. 35: 819–826.
- Zhu TJ, Yang JQ, Tang WQ. 2008. MtDNA control region sequence structure of the genus Coilia in Yangtze River estuary. J Shanghai Fish Univ. 17:152–157.