Abstract
Complete mitochondrial genomes of Isospora amphiboluri and an unnamed Isospora sp. (Eimeriidae, Coccidia, Apicomplexa) were sequenced. These coccidia infect the bearded dragon, Pogona vitticeps (Sauria: Agamidae) and black-throated laughingthrush Garrulax chinensis (Aves: Passeriformes), respectively. Genome organization and gene content were conventional. The circular-mapping mt genomes of I. amphiboluri (6264 bp) and unnamed Isospora sp. (6225 bp) each contained three protein-coding genes (COI, COIII, and CytB), 17 gene fragments encoding large subunit (LSU) rRNA and 14 gene fragments encoding small subunit (SSU) rRNA. Like other apicomplexan parasites, no tRNA was encoded by either genome. The comparable mitochondrial genome sequences and structures of Isospora and Eimeria species confirm the close relationship between these eimeriid genera of apicomplexan parasites.
Members of the genus Isospora (Apicomplexa, Coccidia, Eimeriidae) are coccidian parasites that typically infect the digestive tract of a wide variety of vertebrate hosts. Isosporosis caused by Isospora amphiboluri Cannon Citation1967 is a common intestinal disease in bearded dragons, Pogona vitticeps (Sauria: Agamidae), with significant mortalities (15% or higher) in young lizards (Kim et al. Citation2002). This parasite was first described in Pogona barbata by Cannon (Citation1967) and later reported to infect P. vitticeps by McAllister et al. (Citation1995). Similarly, newly hatched passerine birds are frequently infected by coccidia in the genus Isospora (see Dolnik Citation2002; Schrenzel et al. Citation2005); in some cases, the parasites infect beyond the digestive tract to cause systemic isosporosis (i.e. ‘atoxoplasmosis’) with profligate replication in macrophages (e.g. Aragão Citation1911). This study reports the mt genome sequences and organization of two Isospora species from vastly divergent host species.
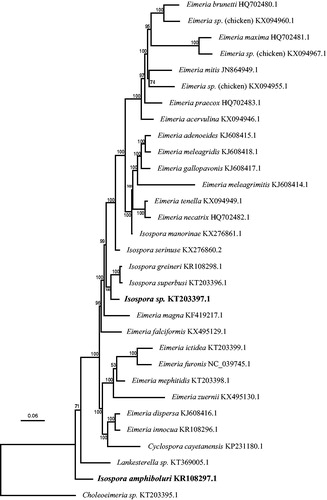
DNA was isolated as described by Hafeez et al. (Citation2014) from faecal oocysts (I. amphiboluri) from a bearded dragon or merozoite-infected tissues of a Black-throated laughingthrush (Garrulax chinensis) that had died of disseminated coccidiosis. The mt genomes were obtained by PCR amplification and amplicon sequencing as described by Ogedengbe et al. (Citation2014). Chromatograms were assembled using the de novo sequence assembly function within Geneious version 6.1.8 or higher (www.geneious.com) to generate a high-quality contig. The newly generated mt sequences were aligned with publically available complete mt genomes from other coccidia to assist in annotation and establish phylogenetic relationships among these parasites ().
The mt genome sequence of I. amphiboluri was 6264 bp in size (GenBank: KR108297.1) and the Isospora sp. of Black-throated laughingthrush was 6225 bp (GenBank: KT203397.1); both were circular-mapping but their physical structure was not determined (i.e. circular or linear concatemer). Similar to mt genomes of related coccidia, both mt genomes possessed three protein-coding genes (COI, COIII, and CytB), 17 gene fragments encoding large subunit (LSU) rRNA and 14 gene fragments encoding small subunit (SSU) rRNA. Fragmented LSU and SSU rDNA is typical for apicomplexan mitochondrial genomes. Gene arrangement and directions were identical to that of other mt genomes from Eimeria spp. (e.g. Ogedengbe et al. Citation2014) and other eimeriid coccidia. The mt genomes of I. amphiboluri and the unnamed Isospora sp were strongly A + T biased overall (66.3 and 65.3%, respectively) with the high A + T bias being maintained in the three CDS (69.4 and 67.8% for CytB; 67.6 and 66.6% for COI; 71.7 and 68.3% for COIII, all respectively).
The availability of complete mt genomes from I. amphiboluri and the unnamed Isospora sp. could provide additional novel DNA markers for molecular epidemiology, population genetics, and for molecular diagnosis of coccidiosis in reptiles and birds.
Disclosure statement
The authors report no conflicts of interests. The authors alone are liable for the content and writing of the manuscript.
Figure 1. Phylogenetic tree based on the protein- and rRNA-coding regions of complete mitochondrial genome sequences from a variety of eimeriid coccidia rooted by a Choleoeimeria species. The unnamed Isospora sp. from the Black-throated laughingthrush was found in a weakly supported clade containing a number of Isospora species infecting passerine birds. Isospora amphiboluri branched as sister taxon to a large clade of eimeriid coccidia The Bayesian analysis was performed on an alignment of 5419 bp from each complete mitochondrial genome using extracted CDS and 33 rDNA fragments; the dataset was partitioned so that the CDS were analysed using a codon-based (mtmet translation) substitution model and the rDNA regions analysed using a GTR + G + I substitution model. Scale bar indicates hypothesized evolutionary divergence and numbers at nodes indicate Bayesian posterior probabilities.
Additional information
Funding
References
- Aragão HB. 1911. Observation sobre algumas hemogregarinas das aves. Mem Inst Oswaldo Cruz. 3:54–64.
- Cannon LRG. 1967. New coccidia from Australian lizards. I. Isospora. Parasitology. 57:227–235.
- Dolnik O. 2002. Some aspects of the biology and host-parasite interactions of Isospora spp. (Protozoa: Coccidiida) of passerine birds [PhD dissertation]. Oldenburg, Germany: Carl von Ossietzky University. http://oops.uni-oldenburg.de/214/97/dolsom03.pdf.
- Hafeez MA, Stasiak I, Delnatte P, El-Sherry S, Smith DA, Barta JR. 2014. Description of two new Isospora species causing visceral coccidiosis in captive superb glossy starlings, Lamprotornis superbus (Aves: Sturnidae). Parasitol Res. 113:3287–3297.
- Kim DY, Mitchell MA, Bauer RW, Poston R, Cho DY. 2002. An outbreak of adenoviral infection in inland bearded dragons (Pogona vitticeps) coinfected with dependovirus and coccidial protozoa (Isospora sp.). J Vet Diagn Invest. 14:332–334.
- McAllister CT, Upton SJ, Jacobson ER, Kopit W. 1995. A description of Isospora amphiboluri (Apicomplexa: Eimeriidae) from the inland bearded dragon, Pogona vitticeps (Sauria: Agamidae). J Parasitol. 81:281–284.
- Ogedengbe ME, El-Sherry S, Whale J, Barta JR. 2014. Complete mitochondrial genome sequences from five Eimeria species (Apicomplexa; Coccidia; Eimeriidae) infecting domestic turkeys. Parasit Vectors. 7:335.
- Schrenzel MD, Maalouf GA, Gaffney PM, Tokarz D, Keener LL, McClure D, Griffey S, McAloose D, Rideout BA. 2005. Molecular characterization of isosporoid coccidia (Isospora and Atoxoplasma spp.) in passerine birds. J Parasitol. 91:635–647.
