Abstract
The complete mitochondrial genome of the leafhopper Chlorotettix nigromaculatus was described in this study. The circular molecule is 15,011 bp in length (GenBank accession no. MK234840), consisted of 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, and a putative control region. The base composition values for the mitogenome were 42.4, 13.3, 10.4, and 33.9% for A, C, G, and T, respectively. All the 22 tRNAs have the typical cloverleaf structures, except for trnS1 which lacked the dihydrouridine arm. All the 13 PCGs initiated with ATN codons and most of them terminated with TAG or TAA codons, whereas cox2, nad1, and nad4 used a single T residue. The rrnL and rrnS genes were 1220 and 745 bp in length, respectively. Phylogenetic analysis using amino acid sequences of 13 PCGs showed that C. nigromaculatus was clustered with six Deltocephalinae species, which was consistent with the conventional classification.
The leafhopper Chlorotettix nigromaculatus belongs to the subfamily Deltocephalinae (Hemiptera: Cicadellidae), which includes 6683 known species around the world (Dai et al. Citation2006). Many leafhopper species in this subfamily are harmful to woody and herbaceous plants through sucking, ovipositing, and virus transmission (Mckamey Citation2002; Du et al. Citation2017). In this study, the C. nigromaculatus samples were collected from Fanjing Mountain in Guizhou Province of China (N2752′, E108°47′). The whole body specimen was preserved in ethanol and stored in the insect specimen room of Guiyang University with an accession number GYU-Col-20180003.
The complete mitogenome of C. nigromaculatus is a circular molecule of 15,011 bp in length (GenBank accession no. MK234840), containing 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rrnL and rrnS), and a putative control region. The gene order and orientation of C. nigromaculatus were identical with other Hemiptera species (Mao et al. Citation2017; Dai et al. Citation2018). The base composition values for C. nigromaculatus mitogenome were 42.4, 13.3, 10.4, and 33.9% for A, C, G, and T, respectively, with an overall A + T content of 76.3%. The AT-skew and GC-skew of this mitogenome were 0.111 and −0.125, respectively. Twenty-four genes were oriented on the majority strand (N-strand), whereas the others were transcribed on the minority strand (J-strand). The C. nigromaculatus mitogenome has a total of 37 bp intergenic spacer sequences, which is made up of 10 regions in the range from 1 to 12 bp. The largest intergenic spacer sequence of 12 bp is located between cox3 and trnG. Gene overlaps were found at eight gene junctions and involved a total of 28 bp, the longest 8 bp overlapping located between trnW and trnC.
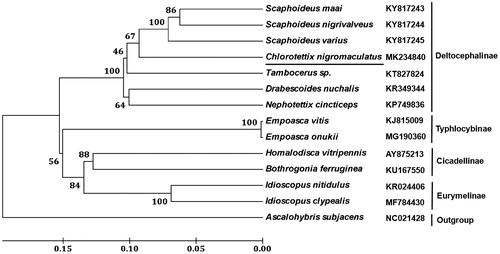
All 13 PCGs initiate with ATN as the start codons (ATG for atp6, cox3, and cob; ATT for nad2 and nad4L; ATA for cox1, cox2, atp8, nad1, nad3, nad4, nad5, and nad6). The conventional termination codons (TAA or TAG) occur in 10 PCGs, and the remaining PCGs including cox2, nad1, and nad4 terminate with a single T residue. The shortest PCG is atp8 gene (153 bp) and the longest one is nad5 gene (1671 bp). The mitogenome harbours the complete set of 22 tRNA genes, ranging from 61 bp (trnH) to 73 bp (trnW). All of the 22 tRNA genes have the typical cloverleaf structure except for the dihydrouridine arm of trnS1 (AGN) forms a simple loop, which is similar to previous report in most animal mitogenomes (Wolstenholme Citation1992). The rrnL gene is located between trnL1 and trnV, with length of 1220 bp and A + T content of 80.25%. The rrnS gene is located between trnV and control region, with a length of 740 bp and A + T content of 76.64%. The control region is located between rrnS and trnI genes is 650 bp in length with an A + T content of 77.2%. Phylogenetic analysis was performed using the neighbour-joining method based on the amino acid sequences of 13 PCGs from C. nigromaculatus and other 12 leafhoppers. The result showed that C. nigromaculatus was clustered with six Deltocephalinae species (), which was consistent with the conventional classification.
Disclosure statement
The authors report no conflicts of interests. The authors alone are responsible for the content and writing of the article.
Additional information
Funding
References
- Dai RH, Li ZZ, Chen XX. 2006. One new genus and species of Euscelinae (Hemiptera, Cicadellidae) from Guizhou, China. Acta Zootaxonomica Sin. 31:592–594.
- Dai RH, Wang JJ, Yang MF. 2018. The complete mitochondrial genome of the leafhopper Idioscopus clypealis (Hemiptera: Cicadellidae: Idiocerinae). Mitochondrial DNA Part B. 3:32–33.
- Du YM, Dai W, Dietrich CH. 2017. Mitochondrial genomic variation and phylogenetic relationships of three groups in the genus Scaphoideus (Hemiptera: Cicadellidae: Deltocephalinae). Sci Rep. 7:16908.
- Mao M, Yang XS, Bennett G. 2017. The complete mitochondrial genome of Macrostelesquadrilineatus (Hemiptera: Cicadellidae). Mitochondrial DNA Part B. 2:173–175.
- Mckamey SH. 2002. The distribution of leafhopper pests in relation to other leafhoppers (Hemiptera; Cicadellidae). Denisia. 176:357–378.
- Wolstenholme DR. 1992. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 141:173–216.