Abstract
Molecular tools have been used in wildlife forensics for species identification of indiscernible samples and to provide evidence against poaching of wildlife and illegal trade of their products. This study was undertaken in identifying a confiscated material that has been seized under the Wildlife (Protection) Act, 1972 of India suspecting that accused has poached a deer species of wild origin for local consumption. The sample was processed for DNA sequencing of the partial fragments of three mitochondrial genes, that is, Cytochrome b, 12S rRNA, and 16S rRNA genes. Surprisingly, generated sequences from the confiscated sample showed 99% homology with Bos indicus, and phylogenetic analysis also clustered the confiscated material with Bos indicus with high bootstrap support. This study upholds the evidence for accused as the suspicion of confiscated material being an illegal product was negated, and the raw meat of cattle was merely for consumption through reliable assessment and validation from a public database. This case study exhibits the value of DNA barcoding and molecular diagnostic techniques to decipher the morphologically altered materials in wildlife forensic cases.
Introduction
Illegal hunting or poaching of wild animals for their products is causing a great loss to biodiversity bringing many species on the verge of extinction (Thakur Citation2014). The illegal trade in flora and fauna offers great financial rewards as compared to the others, which has deeply attracted attention of the criminals to make easy money (Ogden et al. Citation2009). Demand in the international market for wildlife products such as ivory, scales, claws, horns, bones, etc. leads to over-exploitation of wildlife which often lead to local extinction as sadly happened in cases of Western Black Rhino (Largot et al. 2007), Javan Rhino (Streicher et al), and Passenger pigeon (Hume Citation2015). Many law enforcement agencies have been in function and showed great efforts to curb this illegal killing and trafficking of animals by implementing the Wildlife Acts, legitimate to protect the wildlife biodiversity in different parts of the globe. Traditionally diagnostic morphological traits have been used as markers to identify the species, but in degraded or highly processed products where the morphology is often disfigured, species confirmation is not possible using morphological traits (Alacs et al. Citation2010). Molecular markers such as Cytochrome b (Mukesh et al. Citation2013; Kumar et al. Citation2016; Thakur et al. Citation2017), Cytochrome C oxidase subunit I (CoI) (Hebert et al. Citation2003) are ideal markers for species identification as they do not require intact specimens and mutation rates coincide with the rate of species evolution. DNA based studies are used to identify the unknown sample using the mitochondrial genes (Alacs et al. Citation2010; Zhang et al. Citation2015; Rajpoot et al. Citation2018). To confirm the species identity, sequences generated from the unknown sample have been primarily compared with the reference database such as NCBI and percent similarity is measured. Thus, database attributes the most likely species to the unknown sequence based on the similarity indices. A further confirmation of species identity is evaluated by the construction of phylogenetic trees where the sequence of unknown identity is supposed to be clustered along with its nearest neighbour (Ogden et al. Citation2009). DNA based identification with the availability of reference sequences, for example, ‘ForCytb’ (Ahlers et al. Citation2017) for species identification can be significant in the implementation of wildlife acts and laws applicable at regional or at global level (Zhang et al. Citation2015; Kumar et al. Citation2016). Several studies have shown the potential of DNA forensics to hold the criminals and proved important evidence against the wildlife crime (Verma and Singh Citation2002; Wong et al. Citation2004; Wasser et al. Citation2007). Besides nabbing the criminals involved in illegal killing and poaching of wildlife, there have been instances, largely go unreported where innocent people are booked under the false charges of wildlife killing. This often happened unintentionally either due to the wrong information supplied to the law enforcement agencies or due to the distorted morphology of the seized material leading to misidentification of the specimen. Thus, the present study has been unique in its finding as it highlighted the use of DNA forensics in identifying innocence of the accused as the sample was misapprehended by the forest officials due to the wrong information.
Case history
Forest Officials, Raidighi Range, West Bengal confiscated raw meat suspected to be of wildlife origin and sent to Zoological Survey of India (ZSI) for species identification to provide evidences of poaching of wildlife and prosecuting the case for legal hearing in the court of law related to the implication of Wildlife (Protection) Act, 1972 of India. Sample received at ZSI was in a plastic container and weighed around 300 g with absolutely no intact clues of species identity. During the investigation, three accused were arrested and held in police custody on the suspicion that meat is of deer origin, presuming the offence happened in the vicinity of the Sunderban National Park, West Bengal.
Materials and methods
Genomic DNA was extracted from the confiscated raw meat using Qiagen DNeasy Blood and Tissue Kit (Qiagen, Germany) and the quality was checked on 1% agarose gel. PCR was performed to amplify the partial fragments of three mitochondrial gene using the universal primers of Cytochrome b (Verma and Singh Citation2003), 12S rRNA (Kocher et al. Citation1989), and 16s rRNA (Palumbi Citation1991) genes to obtain sequences independently for species-level identification. The PCR amplicon was cleaned up using Exo-SAP treatment and Sanger sequencing was performed on Genetic analyzer 3730 (Applied Biosystems, California, USA) using the Big Dye terminator cycle sequencing kit v3.1 (Applied Biosystems).
Data analysis
The sequences obtained were checked and cleaned manually using Sequencher version 5.4.6 (www.genecode.com). Sequences thus generated were matched for similarity search using the online tool ‘BLAST’ (Basic Local Alignment Sequencing Tool) of NCBI. Most homologous sequences, considering a threshold of ≥88% similarity, were downloaded from NCBI and Multiple Sequence Alignment was done by using ClustalW tool as implemented in MEGA X (Kumar et al. Citation2018). Neighbour-joining (NJ) phylogenetic trees were reconstructed following maximum likelihood with Hasegawa Kishino-Yano model (Cytb and 12SrRNA genes) and Timura 3-parameter model (16S rRNA gene) based on the lowest BIC values using MEGA X (Kumar et al. Citation2018). Genetic distance matrix of all three genes (Cytb, 12S, and 16S gene) was constructed using Nei’s DA model in MEGA X.
Results
Yielded sequence length of the confiscated material for three genes was 428bp (Cytb), 423bp (12S rRNA), and 561bp (16S rRNA) and homology search showed a 99–100% similarity with Bos indicus covering the sequence length 428bp, 423bp, and 561bp of Cytb, 12Sr RNA, and 16S rRNA genes, respectively. In contrast, the percent similarity of confiscated material with any of the deer species taken into analysis was less than 92%. The genetic distance matrix for all three genes revealed that the suspected confiscated material showed absolutely no genetic distance, that is, Nei’s DA 0.00 with the known sequences of domestic cattle, Bos indicus ( ).
Table 1. Nei’s genetic (DA) distance matrix (A) Cytb, (B) 12S rRNA, and (C) 16S rRNA.
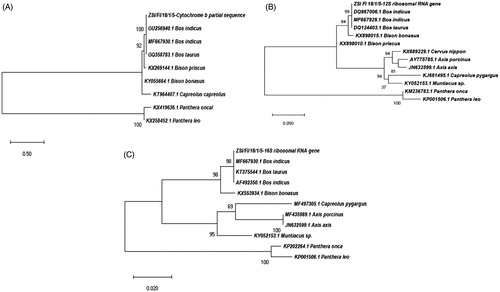
The NJ trees for all three genes also corroborated findings of BLAST and genetic distance indices and also grouped questioned meat sample with domestic cattle, Bos indicus with 100% bootstrap support (). The estimated bootstrap values higher or equal to 70% usually correspond to probabilities higher or equal to 95% which means that the topology is close to real, giving a quantitative measurement of certainty of the assignment of a sample to a particular species. Considering the evidence of percent similarity, genetic distance matrix, and the NJ trees of three genes, it has been concluded that confiscated material was a cattle origin than a deer species.
Figure 1. Reconstruction of the neighbour-joining (NJ) phylogenetic trees using the Maximum Likelihood method and the Hasegawa–Kishino–Yano (Cyt b and 12s r RNA) and Tamura 3-parameter model (16S r RNA) with 1000 bootstrap replicates. (A) Cyt b, (B) 12S rRNA gene, and (C) 16S r RNA genes.
The confiscated material in the form of the tissue stored in alcohol and DNA extract has been archived in the reference repository of wildlife seizures at Centre for DNA Taxonomy of ZSI, Kolkata. The resulted sequences of Cytochrome b, 12Sr RNA, and 16Sr RNA gene were submitted to NCBI/GenBank with the accession no – MH423748, MH423715, MH423743.
Discussion
Many studies have already shown the use of molecular technology and barcoding in wildlife forensics (Verma and Singh, Citation2003; Wong et al. Citation2004; Wasser et al. Citation2007; Kumar et al. Citation2016; Mukesh et al. 2017). This case study has further proven the utility of DNA barcoding in wildlife forensics in identifying the raw meat of cattle origin with no discernible morphology. Interestingly, this case has been unique in generating evidence in support of the accused to nullify the false accusation and assist the court of law in concluding the case. This study has therefore shown the potential that how the accused not guilty by involving in the crime can be identified while he was unknowingly and incorrectly booked under Wildlife (Protection) Act, 1972. We believe this study will encourage and provide new dimensions to the entire law enforcement networks including government agencies, institutes, and court of law in searching all the possible ways in order to generate evidence in cases where morphological identity has been distorted for any reason.
Acknowledgements
The authors thank the Forest Officials, Raidighi Range, West Bengal for providing the case property for species identification.
Disclosure statement
The authors declare that they have no conflict of interest.
Additional information
Funding
References
- Ahlers N, Creecy J, Frankham G, Johnson RN, Kotze A, Linacre A, McEwing R, Mwale M, Rovie-Ryan JJ, Sitam F, Webster LMI. 2017. ‘ForCyt’ DNA database of wildlife species. Forensic Sci Int: Genet Suppl Series. 6:e466–e468.
- Alacs EA, Georges A, FitzSimmons NN, Robertson J. 2010. DNA detective: a review of molecular approaches to wildlife forensics. Forensic Sci Med Pathol. 6:180–194.
- Hebert PDN, Ratnasingham S, DeWaard JR. 2003. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc R Soc Lond Ser B Biol Sci. 270:96–99.
- Hill E, Linacre AMT, Toop S, Murphy NP, Strugnell JM. 2017. The complete mitochondrial genome of Axis porcinus (Mammalia: Cervidae) from Victoria, Australia, using MiSeq sequencing. Mito DNA Part B Resour. 2:453–454.
- Hume JP. 2015. Large-scale live capture of Passenger Pigeons Ectopistes migratorius for sporting purposes: overlooked illustrated documentation. Bull Br Orn Club. 135:174–184.
- Kocher TD, Thomas WK, Meyer A, Edwards SV, Pääbo S, Villablanca FX, Wilson AC. 1989. Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci USA. 86:6196–6200.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Kumar VP, Mukesh RA, Shukla M, Kumar D, Goyal SP. 2016. Illegal trade of Indian Pangolin (Manis crassicaudata): genetic study from scales based on mitochondrial genes. Egypt J Forensic Sci. 6:524–533.
- Lagrot I, Lagrot JF, Bour P. 2007. Probable extinction of the western black rhino, Diceros bicornis longipes: 2006 survey in northern Cameroon. Pachyderm. 43:19–28.
- Martins RF, Fickel J, Le M, van Nguyen T, Nguyen HM, Timmins R, Gan HM, Rovie-Ryan JJ, Lenz D, Förster DW, Wilting A. 2017. Phylogeography of red muntjacs reveals three distinct mitochondrial lineages. BMC Evol Biol. 17:34.
- Mukesh Singh SK, Shukla M, Sharma LK, Mohan N, Goyal SP, Sathyakumar S. 2013. Identification of galliformes through Forensically Informative Nucleotide Sequencing (FINS) and its implication in wildlife forensics. J of Forensic Res. 4:195.
- Ogden R, Dawnay N, McEwing R. 2009. Wildlife DNA forensics — bridging the gap between conservation genetics and law enforcement. Endanger Species Res. 9:179–195.
- Palumbi S. 1991. Simple fool's guide to PCR. Honolulu: University of Hawaii.
- Rajpoot A, Kumar VP, Bahuguna A, Singh T, Joshi S, Kumar D. 2018. Wildlife forensics in battle against veneration frauds in Uttarakhand, India: identification of protected Indian monitor lizard in items available in the local market under the name of Hatha Jodi. Mit DNA Part B. 3:927–934.
- Thakur M. 2014. Role of DNA Forensics in curbing illegal wildlife trade. WWF Newsletter PANDA, Illegal Wildlife Trade in India: Special Issue. 11–12.
- Thakur M, Javed R, Kumar VP, Shukla M, Singh N, Maheshwari A, Mohan N, Wu D-D, Zhang Y-P. 2017. DNA forensics in combating food frauds: a case study from China in identifying canned meat labeled as deer origin. Cur Science. 112:2449–2442.
- Verma SK, Singh L. 2003. Novel universal primers establish identity of an enormous number of animal species for forensic application. Mol Ecol Notes. 3:28–31.
- Wasser SK, Mailand C, Booth R, Mutayoba B, Kisamo E, Clark B, Stephens M. 2007. Using DNA to track the origin of the largest ivory seizure since the 1989 trade ban. Proc Natl Acad Sci USA. 104:4228–4233.
- Wasser SK, Shedlock AM, Comstock K, Ostrander EA, Mutayoba B, Stephens M. 2004. Assigning African elephant DNA to geographic region of origin: applications to the ivory trade. Proc Natl Acad Sci USA. 101:14847–14852.
- Wong KL, Wang J, But PPH, Shaw PC. 2004. Application of cytochrome b DNA sequences for the authentication of endangered snake species. Forensic Sci Int. 139:49–55.
- Zhang H, Miller MP, Yang F, Chan HK, Gaubert P, Ades G, Fischer GA. 2015. Molecular tracing of confiscated pangolin scales for conservation and illegal trade monitoring in Southeast Asia. Glob Ecol Conserv. 4:414–422.
