Abstract
Saprosma merrillii is a plant of the family Rubiaceae. So far, there is no study on the genome of S. merrillii. Here, we report and characterize the complete plastid genome sequence of S. merrillii in an effort to provide genomic resources useful for promoting its conservation. The complete plastome is 155,820 bp in length and contains the typical structure and gene content of angiosperm plastome, including two Inverted Repeat (IR) regions of 26,228 bp, a Large Single-Copy (LSC) region of 85,625 bp and a Small Single-Copy (SSC) region of 17,739 bp. The plastome contains 114 genes, consisting of 80 unique protein-coding genes, 30 unique tRNA genes, and 4 unique rRNA genes. The overall A/T content in the plastome of S. merrillii is 62.50%. The complete plastome sequence of S. merrillii will provide a useful resource for the conservation genetics of this species as well as for the phylogenetic studies for Rubiaceae.
Saprosma merrillii is a plant of the family Rubiaceae. It is a shrub only distributed in Hainan province of China and often grows at sparse forests or forest margin (Tao and Taylor Citation2011). So far, there have been no studies on the genome of S. merrillii. Consequently, the genetic and genomic information is urgently needed to promote its systematics research and the development of conservation value of S. merrillii. Here, we report and characterize the complete plastid genome sequence of S. merrillii (GenBank accession number: MK203879, this study) in an effort to provide genomic resources useful for promoting its conservation.
In this study, S. merrillii was sampled from Jianfeng Mountain (18.74°N, 108.86°E), which is a National Nature Reserve of Hainan, China. A voucher specimen (Wang et al. B66) was deposited in the Herbarium of the Institute of Tropical Agriculture and Forestry (HUTB), Hainan University, Haikou, China.
The experiment procedure is as reported in Zhu et al. (Citation2018). Around 6 Gb clean data were assembled against the plastome of Morinda officinalis (NC_028009.1) (Zhang and Handy Citation2016) using MITO bim v1.8(Le Petit-Quevilly, France) (Hahn et al. Citation2013). The plastome was annotated using Geneious R8.0.2 (Biomatters Ltd., Auckland, New Zealand) against the plastome of M. officinalis (NC_028009.1). The annotation was corrected with DOGMA (Wyman et al. Citation2004).
The plastome of S. merrillii was found to possess a total length 155,820 bp with the typical quadripartite structure of angiosperms, containing two Inverted Repeats (IRs) of 26,228 bp, a Large Single-Copy (LSC) region of 85,625 bp and a Small Single-Copy (SSC) region of 17,739 bp. The plastome contains 114 genes, consisting of 80 unique protein-coding genes, 30 unique tRNA genes, and 4 unique rRNA genes. The overall A/T content in the plastome of S. merrillii is 62.50%, which the corresponding value of the LSC, SSC, and IR region were 64.70, 68.30, and 57.00%, respectively.
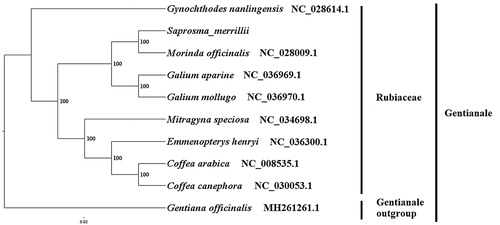
We used RAxML (Stamatakis Citation2006) with 1000 bootstraps under the GTRGAMMAI substitution model to reconstruct a maximum likelihood (ML) phylogeny of eight published complete plastomes of Rubiaceae, using Gentiana officinalis (http://www.efloras.org/florataxon.aspx?flora_id=2&taxon_id=10362">Gentianaceae, Gentianable) as an outgroup. The phylogenetic analysis indicated that S. merrillii is closer to M. officinalis (). Most nodes in the plastome ML trees were strongly supported. The complete plastome sequence of S. merrillii will provide a useful resource for the conservation gengtics of this species as well as for the phylogenetic studies for Rubiaceae.
Figure1. The best ML phylogeny recovered from 10 complete plastome sequences by RAxML. Accession numbers: Saprosma merrillii (MK203879, this study), Emmenopterys henryi NC_036300.1, Galium mollugo NC_036970.1, Mitragyna speciosa NC_034698.1, Gynochthodes nanlingensis NC_028614.1, Morinda officinalis NC_028009.1, Coffea arabica NC_008535.1, Galium aparine NC_036969.1, Coffea canephora NC_030053.1, outgroup: Gentiana officinalis MH261261.1.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Tao C, Taylor CM. 2011. Flora of China. Vol 19. Beijing: Science Press; p. 320–322.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zhang N, Handy SM. Submitted (31-Oct-2016) Center of Food Safety and Nutrition, Foodand Drug Adminstration, 5100 Paint Branch Pkwy, College Park, MD20740, USA. https://www.ncbi.nlm.nih.gov/nuccore/NC_028009.1/
- Zhu ZX, Mu WX, Wang JH, Zhang JR, Zhao KK, Friedman CR, Wang HF. 2018. Complete plastome sequence of Dracaena cambodiana (Asparagaceae): a species considered “Vulnerable” in Southeast Asia. Mitochondrial DNA B Resour. 3:620–621.
