Abstract
The complete mitochondrial genome of Artemia sinica was obtained using the next-generation sequencing (NGS) method. The mitochondrial genome is a circular molecule of 15,689 bp in length, with the typical structure of 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs) and 2 ribosomal RNA genes, and a non-coding control region (CR). The base composition is 31.53% A, 18.99% C, 16.50% G, and 32.98% T, with an A + T content of 64.51%. All tRNAs have a cloverleaf structure excepting tRNA-Ser1, that represents the D-loop structure.
The genus Artemia consists of seven bisexual species and a large number of parthenogenetic populations with different ploidy degrees (Asem et al. Citation2010). So far, the complete mitogenome of three bisexual species of Artemia (Artemia franciscana, Artemia urmiana, and Artemia tibetiana) have already been characterized(Valverde et al. Citation1994; Zhang et al. Citation2013). In this study, we sequenced and described the complete mitochondrial genome of Artemia sinica (GenBank: MK069595). The present study is a part of a more comprehensive project of characterizing the complete mitochondrial genome of genus Artemia.
The cysts of A. sinica (ARC: 1166) were collected from Ejinor Lake (Inner Mongolia, China) and stored in the Artemia Reference Center (Ghent University, Belgium). The total DNA was extracted from a cultured adult specimen. A genomic library was established followed by paired-end (2 × 150 bp) next-generation sequencing (10 Gb), using the Illumina HiSeq X-ten sequencing platform. Quality checks for sequencing reads were performed by FastQC (Andrews Citation2010) and the sequences were assembled and mapped to the reference Artemia mitochondrial genome (A. franciscana, X69067) with Spades v3.9.0 (Bankevich et al. Citation2012) and bowtie v2.2.9 (Langmead and Salzberg Citation2012). Putative tRNA genes were determined using the tRNAscan-SE2.0 (http://lowelab.ucsc.edu/tRNAscan-SE/) and ARWEN (http://130.235.46.10/ARWEN/) online software. All genes were annotated based on gene order on the reference mitochondrial map and using BLAST analysis (https://blast.ncbi.nlm.nih.gov). Additionally, to annotate PCGs and tRNAs, the position of start and stop codons, and secondary structures and the position of anticodons were re-considered, respectively.
The complete mitogenome of A. sinica was 15,689 bp in length, with 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), 2 ribosomal RNAs (rRNAs), and a control region (CR). The overall nucleotide composition of the major strand of the A. sinica mitogenome was as follows: 31.53% A, 18.99% C, 16.50% G, and 32.98% T, with a total A + T content of 64.51%.
Nine tRNAs (tRNA-Ile, tRNA-Gln, tRNA-Cys, tRNA-Tyr, tRNA-Phe, tRNA-His, tRNA-Pro, tRNA-Leu, and tRNA-Val) and four PCGs (ND5, ND4, ND4L, and ND1), as well as both rRNAs were encoded on the light strand. Just six PCGs (ND2, COX1, ATP6, COX3, CYTB, and ND1) began with the common ATG start codon. Stop codons included TAA (ND2, ATP8, ATP6, ND3, ND4L, CYTB, and ND1), TAG (ND6) and non-complete codons T (COX1, COX2, COX3 and ND5, and ND4). The 12S rRNA and 16S rRNA were separated by the tRNA-Val.
Based on the results, the highest and lowest values of %GC tRNA composition belong to tRNA-Lys (46.9%) and tRNA-Glu (18.2%), respectively. Secondary structures of tRNA-Ser1 showed the D-loop structure. The longest and shortest tRNAs were tRNA-Ser2 (67 bp) and tRNA-Ala (59 bp), respectively.
CR was located between 12S rRNA and tRNA-Met, with an A + T content of 67.58%. The longest gap and overlapping were determined between tRNA-Gln/tRNA-Cys (54 bp) and tRNA-Phe/ND5 (13 bp), respectively.
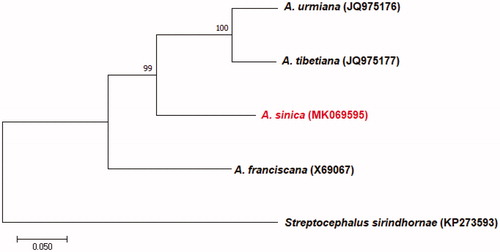
The phylogenetic relationship of A. sinica with members of the genus Artemia was determined from a concatenated dataset including the 13 PCGs and 2 rRNAs using the software MEGA 7.0.26 v. (Kumar et al. Citation2016) with 1000 bootstrap replicates and a GTR model (). According to the result, A. sinica was placed as a clade sister to other Asian spp. All Asian species were clearly separated from American A. franciscana.
Figure 1. Phylogenetic tree showing the relationship among A. sinica and three other species from the Artemia based on maximum-likelihood (ML) approach. Numbers behind each node denote the bootstrap support values. The GenBank accession numbers are indicated on the right side of species names. Streptocephalus sirindhornae was used as an outgroup.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Andrews S. 2010. FastQC: A quality control tool for high throughput sequence data. [accessed 2018 Oct 4]. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- Asem A, Rastegar-Pouyani N, De los Rios P. 2010. The genus Artemia Leach, 1819 (Crustacea: Branchiopoda): true and false taxonomical descriptions. Lat Am J Aquat Res. 38:501–506.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Bio Evol. 33:1870–1874.
- Langmead B, Salzberg S. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359.
- Valverde J, Batuecas B, Moratilla C, Marco R, Garesse R. 1994. The complete mitochondrial DNA sequence of the Crustacean Artemia franciscana. J Mol Evol. 39:400–408.
- Zhang, H, Luo Q, Sun J, Liu F, Wu G, Yu J, Wang W. 2013. Mitochondrial genome sequences of Artemia tibetiana and Artemia urmiana: assessing molecular changes for high plateau adaptation. Sci China Life Sci. 56: 440–452.
