Abstract
American ginseng, Panax quinquefolius L. is one of the most widely used and perennial understory herbal remedies in the world, belongs to the Araliaceae family. It is native to Canada, Eastern United States, and North America. American ginseng has always been known as ‘green gold’ in North America. In this study, the complete chloroplast genome of P. quinquefolius L. was determined by the BGISEQ-500 Sequencing. The complete chloroplast genome of American ginseng, (P. quinquefolius L.) was 156,267 bp in length and displays a typical quadripartite structure of the large (LSC, 86,124 bp) and small (SSC, 18,069 bp) single-copy regions, separated by a pair of inverted repeat regions (IRs, 26,037 bp each). It harbours 133 functional genes, including 89 protein-coding genes, 36 transfer RNA, and 8 ribosomal RNA genes species. The overall nucleotide composition was: 30.6% A, 31.4% T, 19.4% C, and 18.6% G, with a total G + C content of 38.0%. Phylogenetic relationship analysis shows that P. quinquefolius L. closely related to Panax ginseng.
American ginseng, Panax quinquefolius L. has been recommended for years as a traditional medicine in America, Canada, North America, China, and East Asia (Cruse-Sanders and Hamrick Citation2004). The major bioactive components of American ginseng are the triterpene saponins known as ginsenosides. American ginseng contains Rb1, Rd, and Re ginsenoside higher than Panax ginseng and Ro ginsenoside is only found saponin in American ginseng (Asafu-Adjaye and Wong Citation2003). It also has been used as a medicinal herb for a multitude of diseases such as cancer, diabetes, cardiovascular ills, and obesity (Wu et al. Citation2013). In this study, the complete chloroplast of American ginseng, P. quinquefolius L. was determined and the phylogenetic relationship analysis was carried out, which contributes to the further research and contribute to the conservation of medicinal plants valuable genetic resource
The specimen of P. quinquefolius L. was isolated from Jilin Agricultural University D06 test field in Changchun, Jilin, China (125.40E; 43.82N) and the DNA of P. quinquefolius L. was stored in Jilin Agricultural University College of Life Science (No. JLAUCLS8). The American ginseng DNA sample was sequenced using the BGISEQ-500 Sequencing (BIG, Shenzhen, CA, CHN). The American ginseng complete chloroplast genome was preliminarily annotated using by BLAST and DOGMA online program (Wyman et al. Citation2004), with default settings to identify protein-coding genes, rRNAs, and transfer RNAs (tRNAs) based on the Plant Plastid Code and BLAST homology searches. The secondary structures of tRNA genes were identified by using the ARAGORN (Laslett and Canback Citation2004) or through manually visual inspection.
The chloroplast genome sequence of P. quinquefolius L. is a closed-circular molecule of 156,267 bp in length, containing a large single copy region (LSC) of 86,124 bp, a small single copy region (SSC) of 18,069 bp, and a pair of inverted repeat regions (IRs) of 26,037 bp. In total 133 genes were annotated on this chloroplast genome, including 89 protein-coding genes (PCG), 36 tRNA genes, and 8 ribosomal RNA genes (rRNA). In the IR regions, a total of 16 genes were found duplicated, including 5 PCG species (rps19, rpl2, rpl23, rps7, and rps12), 7 tRNA species (trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, and trnN-GUU), and 4 rRNA species (rrn16, rrn23, rrn4.5, and rrn5). The overall nucleotide composition is: 30.6% A, 31.4% T, 19.4% C, and 18.6% G, with a total G + C content of 38.0%.The annotated chloroplast genome was submitted to GenBank database under accession No.MH056423.
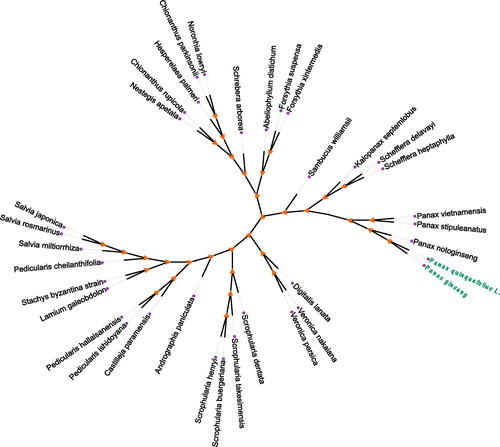
For the phylogenetic relationship, we selected P. quinquefolius L. and other 34 plants chloroplast genomes from GenBank to assess between them. The genome-wide alignment of all plant complete chloroplast genomes was done by HomBlocks (Bi et al. Citation2018). The phylogenetic trees were reconstructed using Maximum-likelihood (ML) methods. ML phylogenetic tree was constructed using MEGA-X with 5000 bootstrap replicate (Kumar et al. Citation2018). All the nodes were inferred with strong support by the ML methods. As shown in the phylogenetic tree (), the chloroplast genome of P. quinquefolius L. showed the closest with P. ginseng.
Figure 1. Phylogenetic tree of Panax quinquefolius L. and other 34 plant complete chloroplast genomes which yielded by Maximum-likelihood analysis. The phylogenetic tree was drawn without setting outgroup. All nodes exhibit above 90% bootstraps. The length of branch represents the divergence distance. The NCBI database accession number of Panax quinquefolius L. to Panax ginseng in the counterclockwise direction is KP036468.1, KX247147.1, KU059178.1, KT748629.1, KC456166.1, KC456167.1, KX510276.1, MG255756.1, MF579702.1, KT274029.1, MG255767.1, MG255759.1, MG255752.1, LN515489.1, MG255753.1, MG255758.1, KY646163.1, KR232566.1, HF586694.1, KY751712.1, KU724141.1, KY562590.1, MG770330.1, KU170194.1, KT959111.1, KF150644.2, MF861203.1, KP718626.1, KP718628.1, MF861202.1, KT724052.1, KT633216.1, KY085895.1, KM067389.1.
Disclosure statement
The authors have declared that no competing interests exist.
Additional information
Funding
References
- Asafu-Adjaye EB, Wong SK. 2003. Determination of ginsenosides (ginseng saponins) in dry root powder from Panax ginseng, Panax quinquefolius, and selected commercial products by liquid chromatography: interlaboratory study. J AOAC Int. 86:1112–1123.
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110:18–22.
- Cruse-Sanders JM, Hamrick JL. 2004. Genetic diversity in harvested and protected populations of wild American ginseng, Panax quinquefolius L. (Araliaceae). Am J Bot. 91:540–548.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32:11–16.
- Wu D, Austin RS, Zhou S, Brown D. 2013. The root transcriptome for North American ginseng assembled and profiled across seasonal development. BMC Genomics. 14:564.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
