Abstract
In this study, the complete mitochondrial genome (mitogenome) sequence of Latreillia valida was determined. The circle genome was 15,097 bp in length and contains 13 protein-coding genes (PCGs), 22 transfer (tRNA) genes, 2 ribosomal (rRNA) genes, and 1 control region. The overall nucleotide composition of this mitogenome was 38.6% for A, 16.0% for C, 9.8% for G, and 35.6% for T and showed 87% identity to Moloha majora. Phylogenetic analysis showed that L. valida was clustered with Homolidae family, suggesting that Latreilliidae and Homolidae had a close relationship.
The banded arrow crab, Latreillia valida is a Brachyura crab in the Latreilliidae family. It inhabits in benthic zone (depth range 30–731 m) of Indo-West Pacific (Castro et al. Citation2003). In the present study, the complete mitochondrial genome sequence of L. valida was first determined.
The specimen of L. valida was collected from Taiwan and deposited at the National Taiwan Ocean University, Keelung, and its genomic DNA was extracted from muscle using the standard phenol/chloroform protocol (Sambrook and Russell Citation2001). Fourteen pairs of primers were designed for sequencing based on the alignment of several Brachyura mitogenomes sequenced in their entirety, using the primer 5 software (Singh et al. Citation1998).
The complete mitochondrial genome of L. valida was 15,097 bp in length (GenBank accession no. MK204361). The total nucleotide composition of this mitogenome is 38.6% for A, 16.0% for C, 9.8% for G, and 35.6% for T. The AT content (74.2%) was higher than GC content (25.8%) which is similar to other brachyuran crabs (Shi et al. Citation2015). The circle genome contains 13 protein-coding genes (PCGs), 22 transfer (tRNA) genes, 2 ribosomal (rRNA) genes, and a control region. Of these, 23 genes were encoded on the heavy strand, and 14 genes were encoded on the light strand. All the PCGs are initiated with ATG codon, except for the ATP6 with ATC. Eleven PCGs have typical stop codon TAA, while COIII and ND5 have the incomplete stop codon T–, respectively. The incomplete stop codons are presumably completed as TAA via posttranscriptional polyadenylation (Ojala et al. Citation1981).
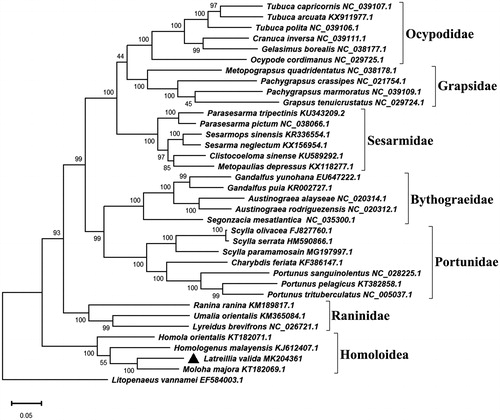
To assess the phylogenetic position of L. valida, we constructed a Maximum-likelihood tree (1000 bootstrap replicates, Mega X software) containing the complete mitochondrial genome of 36 decapod species (). Phylogenetic analysis showed that all the Brachyura crabs were clustered into a group. Latreillia valida was clustered with three Homolidae crabs and all the four species belong to the superfamily of Homoloidea, of which L. valida showed a closest genetic relationship to Moloha majora. Phylogenetic analysis of the brachyuran crabs based on small subunit nuclear ribosomal RNA also recognized Latreilliidae was nested within the Homolidae (Ahyong et al. Citation2007). These findings suggest the close relationship between Latreilliidae and Homolidae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Ahyong ST, Lai JC, Sharkey D, Colgan DJ, Ng PK. 2007. Phylogenetics of the brachyuran crabs (Crustacea: Decapoda): the status of Podotremata based on small subunit nuclear ribosomal RNA. Mol Phylogenet Evol. 45:576–586.
- Castro P, Williams AB, Cooper LL. 2003. Revision of the family Latreilliidae Stimpson, 1858 (Crustacea, Decapoda, Brachyura). Zoosystema. 25:601–634.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290:470.
- Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. 3rd ed. New York: Cold Spring Harbor Laboratory Press.
- Shi G, Cui Z, Hui M, Liu Y, Chan TY, Song C. 2015. The complete mitochondrial genomes of Umalia orientalis and Lyreidus brevifrons: the phylogenetic position of the family Raninidae within Brachyuran crabs. Mar Genom. 21:53–61.
- Singh VK, Mangalam AK, Dwivedi S, Naik S. 1998. Primer premier: program for design of degenerate primers from a protein sequence. Biotechniques. 24:318–319.