Abstract
Castanea seguinii is an endemic chestnut species in China, which is widely distributed over central and southeastern China. The chloroplast genome sequences were finally resolved by high-throughput sequencing platform Illumina Hiseq X Ten. Totally the genome is 160,419 bp in size, AT rich (63.3%), comprising a couple of 25,694 bp long inverted repeat regions, separated by a 90,055 bp large and 18,976 bp small single copy regions. 79 protein-coding genes, four ribosomal RNA genes and 30 transfer RNA genes were identified and annotated. The results of the molecular phylogenetic analysis exhibited that C. seguinii showed the closest relationship with C. mollissima, with high bootstrap support.
Castanea seguinii, an endemic species in China, is widely distributed in 14 provinces over central and southeastern China (Lang and Huang Citation1999). Its natural habitats include mountains and hills where the altitude is between 500 and 1000 m (Qi and Tang Citation2005). C. seguinii blooms normally two or three times a year and produces high fruit yield. Its wood is hard, durable, and most resistant to decay (Shen Citation2015). Unfortunately, the population of C. seguinii decreased sharply due to the ecological damage caused by human activities in the last few decades (Huang Citation1998). Hence, urgent actions are needed to protect and make good use of C. seguinii, the germplasm resources. The goal of the study was to report the chloroplast genome sequences of C. seguinii, and provide information for the study of genetic diversity and phylogenetic relationship. The C. seguinii chloroplast genome annotated has been deposited at GenBank public database under the accession MK248944.
Fresh young leaves of C. seguinii were harvested from an individual at Dawushan, Fengshan, Luotian, Hubei, China (30°57′39.04″N, 115°27′52.48″E). The voucher specimen (LT_2017_SDIP) was deposited in an ultra-low temperature freezer in Shandong Institute of Pomology, Taian, China. Total genomic DNA was isolated from liquid-nitrogen-freezed leaves with CTAB method (Doyle and Doyle Citation1987). Purified total genomic DNA was used for constructing library and sequencing with the paired-end sequencing method on Illumina Hiseq X Ten platform. Raw reads (2.6 G) were filtered to generate clean reads with trim_galore v0.4.4. Then the filtered reads were aligned to the chloroplast genome of C. mollissima (GenBank accession KY951992) (Cheng et al. Citation2018) as a reference using mitoMaker v1.14. All of the genes were annotated using the program GeSeq by comparison with the chloroplast genome of C. mollissima.
The chloroplast genome of C. seguinii is circular-mapping molecule of 160,419 bp in size. It is a typical quadripartite structure containing two inverted repeat regions of 25,694 bp each, a 90,055 bp large and 18,976 bp small single copy regions. The total AT content of C. seguinii chloroplast genome is 63.3%. The AT contents of the large and small single copy regions, 65.4 and 69.1%, respectively, are obviously higher than that of inverted repeat regions 57.2%.
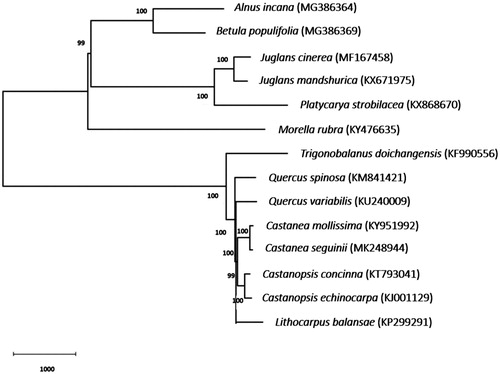
The chloroplast genome of C. seguinii contains 79 protein-coding genes, 30 transfer RNA genes, and 4 ribosomal RNA genes. To investigate the molecular phylogenetic relationships, 14 complete chloroplast genomes were selected to construct a neighbour-joining tree using MEGA software version 6.0 and bootstrap analysis of 1000 replicates. The five species in Fagaceae were clustered together (). C. seguinii and C. henryi formed a clade. Castanea were sisters to Castanopsis with a 99% bootstrap support value and then clustered with Lithocarpus, Quercus and Trigonobalarus.
Disclosure statement
The authors declare that they have no conflict of interest.
Additional information
Funding
References
- Cheng LL, Huang WG, Lan YP, Cao QC, Su SC, Zhou ZJ, Wang JB, Liu JL, Hu GL. 2018. The complete chloroplast genome sequence of the wild Chinese chestnut (Castanea mollissima). Conservation Genet Resour. 10:291–294.
- Doyle JJ, Doyle JD. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang HW. 1998. Review of current research of the world Castanea species and importance of germplasm conservation of China native Castanea species. J Wuhan Bot Res. 16:171–176.
- Lang P, Huang HW. 1999. Genetic diversity and geographic variation in natural populations of the endemic Castanea species in China. Bot Sinica. 41:651–657.
- Qi CJ, Tang GG. 2005. Tree sciences. Beijing (China): Forestry Press; p. 286–287.
- Shen GN. 2015. Science and practice of fruit trees in China. Xian (China): Shaanxi Science and Technology Press; p. 23–25.