Abstract
Based on previous studies of Marchantia polymorpha subsp. ruderalis chloroplast genomes, few sequence variations were identified. Here, we sequenced complete mitochondrial genome of Korean isolate, named as KDBI00084, for uncovering sequence variations on mitochondrial genomes. Its length is 186,196 bp and it contains 99 genes (67 protein-coding genes, 3 rRNAs, and 29 tRNAs). The overall GC content is 42.6%. Only seven single nucleotide polymorphisms are identified grouped by two regions. Each region is covered by inverted repeats. It is valuable to understand the reason why mitochondrial genomes of M. polymorpha subsp. ruderalis have few sequence variations as same as chloroplast genome between Korean and Japanese isolates.
Complete mitochondrial genome of Marchantia polymorpha was published in 1992 as a first mitochondrial genome of Bryophytes (M68929; Oda et al. Citation1992). Consequently, additional researches of this have presented that it might not be from M. polymorpha but Marchantia paleacea (Kisiel et al. Citation2011; Kijak et al. Citation2013; Crandall‐Stotler et al. Citation2016; Bowman et al. Citation2017). Based on resequencing project of Korean M. polymorpha subsp. ruderalis (Korean isolate; KBDI00084; InfoBoss Co., Ltd.), we completed its chloroplast genome (Kwon et al., in submission), presenting only four single nucleotide polymorphisms (SNPs). Variation ratio (0.0033%) is too low in comparison to that of the whole genome of Korean isolate (467,405 SNPs against 205Mb genomes; 0.23%). To confirm whether mitochondrial genome of M. polymorpha subsp. ruderalis has enough variations or not, we completed whole mitochondrial genome of Korean M. polymorpha subsp. ruderalis named as KBDI00084.
M. polymorpha subsp. ruderalis was isolated in Mt. Cheonma, Korea [Voucher in InfoBoss Cyber Herbarium (IN); Kwon W., IB-50002]. Total DNA was extracted from fresh thalli by using a DNeasy Plant Mini kit (QIAGEN, Hilden, Germany) and was sequenced by HiSeq4000 (Macrogen Inc., Seoul, Korea). de novo assembly was done by Velvet 1.2.10 (Zerbino and Birney Citation2008) and confirmed by alignment using BWA 0.7.17 (Li Citation2013) and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.1.5 (Biomatters Ltd, Auckland, New Zealand) was used for annotation with M. polymorpha subsp. ruderalis Kitashirakawa-2 mitochondrial genome (reference genome; NC_037508).
Mitochondrial genome of Korean M. polymorpha subsp. ruderalis (KBDI00084; Genbank accession is MK202951) is 186,196 bp, same to the reference genome (Bowman et al. Citation2017). Its GC ratio is 42.6%, similar to that of M. paleacea (42.4%) and it contains 99 genes (67 protein-coding genes, 3 rRNAs, and 29 tRNAs), which is smaller than that of M. paleacea (110 genes).
We identified seven SNPs on mitochondrial genome of M. polymorpha against reference genome isolated in Japan (NC_037508). Interestingly, these seven SNPs are found in two regions surrounded by inverted repeats (IRs): (i) four SNPs surrounded by 16 bp IR (genomic coordination is 29,639–29,654 and 29,659–29,674) and (ii) three SNPs surrounded by 14 bp IR (179,936–179,949 and 179,953–179,966). Moreover, four and three SNPs from KBDI00084 are inverse sequences of those of reference genome. In addition, mitochondrial genome of another strain, Takaragaike-1, of M. polymorpha subsp. ruderalis was also assembled confirming that there is no sequence variation against the reference mitochondrial genome.
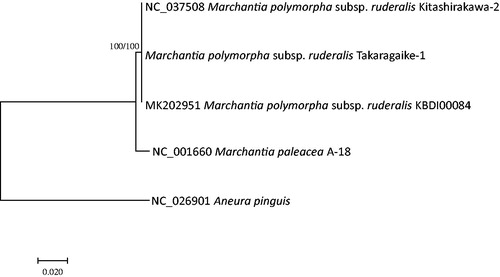
Four complete mitochondrial genomes of class Marchantiopsida including three M. polymorpha subsp. ruderalis and Aneura pinguis (Myszczyński et al. Citation2017) from class Jungermanniopsida were analyzed for constructing phylogenetic trees using both maximum likelihood (bootstrap repeat is 1,000) and neighbor joining (bootstrap repeat is 10,000) methods. Alignment was conducted by MAFFT 7.388 (Katoh and Standley Citation2013) and phylogenetic trees were calculated by MEGA X (Kumar et al. Citation2018). The trees reflect few sequence variations on mitochondrial genome of Marchantia polymorpha subsp. ruderalis in comparison to that of M. paleacea (). It is valuable to understand the reason why mitochondrial genomes of M. polymorpha subsp. ruderalis have few sequence variations as same as its chloroplast genomes between Korean and Japanese isolates.
Figure 1. Maximum likelihood (bootstrap repeat is 1,000) and neighbor joining (bootstrap repeat is 10,000) phylogenetic trees of liverworts based on five complete mitochondrial genomes: M. polymorpha subsp. ruderalis (MK202951, NC_037508, and assembled sequence from Takaragaike-1 strain), M. paleacea (NC_001660), and Aneura pinguis (NC_026901). The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor joining trees, respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bowman JL, Kohchi T, Yamato KT, Jenkins J, Shu S, Ishizaki K, Yamaoka S, Nishihama R, Nakamura Y, Berger F. 2017. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell. 171(2):287–304.e215.
- Crandall-Stotler BJ, Hart ML, Long DG, Forrest LL. 2016. Divergence times and the evolution of morphological complexity in an early land plant lineage (Marchantiopsida) with a slow molecular rate. New Phytologist. 209(4):1734–1746.
- KatohK, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kijak H, Kisiel K, Miwa H, Cieślewicz A, Odrzykoski IK. 2013. What has actually been sequenced? The organellar genome sequences of Marchantia polymorpha from NCBI GeneBank fit M. paleacea subsp. diptera rather than M. polymorpha sensu lato. IAB Conference, The Natural History Museum, London, UK.
- Kisel K, Miwa H, Odrzykoski IJ. 2011. Taxonomic identification of chloroplast genome of Marchantia polymorpha using DNA barcode sequences. In Fourth International Barcode of Life Conference.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 35(6):1547–1549.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:1303.3997. https://doi.org/10.1080/23802359.2018.1551091
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Myszczyński K, Bączkiewicz A, Buczkowska K, Ślipiko M, Szczecińska M, Sawicki J. 2017. The extraordinary variation of the organellar genomes of the Aneura pinguis revealed advanced cryptic speciation of the early land plants. Sci Rep. 7(1):9804.
- Oda K, Yamato K, Ohta E, Nakamura Y, Takemura M, Nozato N, Akashi K, Kanegae T, Ogura Y, Kohchi T. 1992. Gene organization deduced from the complete sequence of liverwort Marchantia polymorpha mitochondrial DNA: a primitive form of plant mitochondrial genome. J Mol Biol. 223(1):1–7.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
