Abstract
The chloroplast genome sequences of Carrierea calycina, endemic in China, is been presented in this article. The species was introduced into the west in the 19th century as a rare ornamental plant. The complete chloroplast genome was 158,327 bp in length with 36.8% of GC content, containing a large single copy region (LSC) of 84,943 bp and a small single copy region (SSC) of 17,570 bp, which were separated by a pair of 27,907 bp inverted repeat regions (IRs). The sequence contained 111 unique genes, including 30 tRNA, 4 rRNA, and 77 protein-coding genes. The phylogenetic position of genus Carrierea has been clarified through Bayesian inference tree based on the chloroplast genome of 14 species.
Keywords:
Carrierea calycina Franch, a species endemic to China, is known as the Goat Horn Tree because of its horn-shaped, green seed pods in China. The genus Carrierea had always been considered in Flacourtiaceae, while it as a relative group of the genera of Salix and Populus was placed into Salicaceae through the molecular study recently (Chase et al. Citation2002). It has recently become known in the west as one of the oldest and rarest ornamental plants, thanks to a tree in Northern Ireland that bloomed nearly a hundred years later. In this study, we first reported the complete chloroplast genome of C. calycina.
The sample of Carrierea calycina was collected in Baoxing, Sichuan (N 30°25′04″, E 102°39′41″, and its voucher specimen (with collection numbers of S. Liao PH20120707-15) has been deposited in the Herbarium of Beijing Forestry University (BJFC). Total genomic DNA was extracted by using modified CTAB method (Doyle and Doyle Citation1987). About 3 Gb high quality, 2 × 150 base pairs PE reads were obtained from High-throughput sequencing, which was performed on Illumina HiSeq 4000 platform at Novogene (http://www.novogene.com, China). We screened out chloroplast reads with the chloroplast genome of its relative Idesia polycarpa (GenBank accession no. KX229742) chloroplast genomes as the reference and used these filtered chloroplast reads for de novo assembly with Geneious R11 (Kearse et al. Citation2012). Plann (Huang and Cronk Citation2015) was used to annotate the assembled chloroplast genome by referring to the relative group, and we perfected the annotation by Geneious.
The chloroplast genome has a total length of 158,327 bp, including a large single copy region (LSC) of 84,943 bp, a small single copy region (SSC) of 17,570 bp and a pair of 27,907 bp inverted repeat regions (IRs). The overall GC-content of the whole plastome genome is 36.8%. The sequence contained 111 unique genes, including 30 tRNA, 4 rRNA, and 77 protein-coding genes.
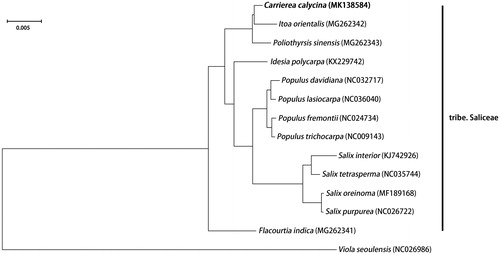
To clarify its phylogenetic position in the Salicaceae, a Bayesian inference (BI) tree based on complete chloroplast genome sequences of 12 other Salicaceae species and Viola seoulensis as the outgroup from NCBI was reconstructed in MrBayes 3.2.3 (Ronquist and Huelsenbeck Citation2003). All the sequences were aligned by MAFFT v6.833 (Katoh et al. Citation2005), and the appropriate nucleotide substitution model for each sequence was according to the Akaike information criterion (AIC; Posada and Buckley Citation2004) by using jModeltest (Posada Citation2008). The posterior probability (PP) of all branch nodes in this phylogenetic tree () is 1.00. As shown in , our research supports that the genera Carrierea, Itoa and Poliothysis are clustered into one clade, which is sister to the clade of the genera Idesia, Populus, and Salix (Alford Citation2005; Zhang et al. Citation2018). Meanwhile, our study further clarifies the phylogenetic relationship among the genera Poliothysis, Itoa, and Carrierea.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Alford MH. 2005. Systematic studies in Flacourtiaceae [dissertation]. Ithaca (NY): Cornell University.
- Chase MW, Zmarzty S, Lledó MD, Wurdack KJ, Swensen SM, Fay MF. 2002. When in doubt, put it in Flacourtiaceae: a molecular phylogenetic analysis based on plastid rbcL DNA sequences. Kew Bull. 57:141–181.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang DI, Cronk QC. 2015. Plann: a command-line application for annotating plastome sequences. Appl Pl Sci. 3:1500026.
- Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511–518.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Posada D, Buckley TR. 2004. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol. 53:793–808.
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25:1253–1256.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Zhang L, Xi Z, Wang M, Guo X, Ma T. 2018. Plastome phylogeny and lineage diversification of Salicaceae with focus on poplars and willows. Ecol Evol. 8:7817–7823.