Abstract
The great spotted woodpecker Dendrocopos major (Aves, Piciformes, Picidae) are the widespread woodpeckers from Eurasia and Northern Africa. The complete mitochondrial genome sequence consisted of 16,845 base pairs (bp) encoding 13 protein-coding genes (PCGs), 2 ribosomal RNAs (rRNAs), 22 transfer RNAs (tRNAs), and 2 non-coding regions (D-loop 1, D-loop 2). The overall base composition of D. major was G + C: 47.5%, A + T: 52.5%, apparently with a slight AT bias. Phylogenetic analysis showed that D. major was closely related to Dendrocopos leucotos.
The great spotted woodpecker Dendrocopos major (Aves, Piciformes, Picidae) are the widespread woodpeckers from Eurasia and Northern Africa. Previously, the genus Dendrocopos included about 25 species. Recently, the number of species in Dendrocopos has reduced to 12 or 13 from the molecular phylogenetic analysis. Despite the changes in the classification of species, there is only one mitogenome paper on the whole mitochondrial genome of this genus. The carcass of D. major was collected at Palgongsan Mountain (36.055540, 128.699755) in Korea. This specimen was deposited in the Institute of Ornithology, Kyungpook National University, Daegu, Republic of Korea.
In the present study, the mitogenome of D. major was completely sequenced and submitted to GenBank: (accession number: KT350609). The sequence was compared with a previously reported mitogenome sequence of Dendrocopos leucotos. The mitochondrial DNA was amplified into two overlapping segments by a long-range PCR method (Koh et al. Citation2018). DNA shotgun sequencing was operated by an Ion PGM™ system (Life Technologies, Gaithersburg, MD, USA) and genome assembly was carried out using the CLC Genomics Workbench 7.5 program (CLC Bio, Aarhus, Denmark ) (Park et al. Citation2018).
The mitogenome was 16,845 bp in size, consisting of 13 protein-coding genes (PCGs), 2 ribosomal RNAs (rRNAs) genes, 22 transfer RNAs (tRNAs), and 2 non-coding regions (D-loops). All the PCGs (ND1, ND2, CO2, ATP8, ATP6, CO3, ND4L, ND4, CytB, ND6) shared start codon ‘ATG’, except for CO1, ND5 (start codon ‘GTG’) and ND3 (start codon ‘ATA’). Regarding the stop codons, 7 PCGs (CO2, ATP8, ATP6, ND3, ND4L, ND4, CytB) shared stop codon ‘TAA’, 3 PCGs (ND1, CO1, ND5) shared stop codon ‘AGG’, ND2 terminated with the incomplete stop codon ‘TA’, CO3 terminated with the incomplete stop codon ‘T’, and ND6 terminated with ‘TAG’. The two rRNA genes were 12S rRNA (960 bases) and 16S rRNA (1592 bases). The first D-loop (1219 bases) was located between tRNA-Thr and tRNA-Pro and the second D-loop (69 bases) was located between tRNA-Glu and tRNA-Phe. The overall base composition of D. major was G: 13.7%, C: 33.8%, A: 28.5%, T: 24.1%, apparently with a slight AT bias (G + C: 47.5%, A + T: 52.5%).
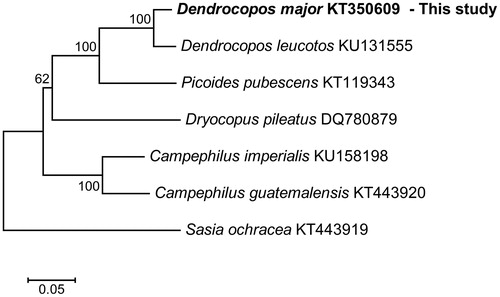
We compared the results from the present study to previous mitogenome research on the family Picidae (Gibb et al. Citation2007; Fuchs et al. Citation2016; Zhang et al. Citation2016; Anmarkrud and Lifjeld Citation2017; Eo Citation2017). Phylogenetic analysis based on the mitogenome sequences using the maximum likelihood general time reversible model (GTR) with gamma distributed (G) plus invariant sites (I) method showed that the D. major was closely related to Dendrocopos leucotos (). Thus, the D. major mitogenome sequence can contribute to phylogenetic knowledge of the genus Dendrocopos and expand the basis of the thrush species.
Disclosure statement
The authors report no conflict of interest. The authors take complete responsibility for the content and writing of this article.
References
- Anmarkrud JA, Lifjeld JT. 2017. Complete mitochondrial genomes of eleven extinct or possibly extinct bird species. Mol Ecol Resour. 17:334–341.
- Eo SH. 2017. Complete mitochondrial genome of white-backed woodpecker Dendrocopos leucotos (Piciformes: Picidae) and its phylogenetic position. Mitochondrial DNA B. 2:451–452.
- Fuchs J, Pons JM, Pasquet E, Bonillo C. 2016. Complete mitochondrial genomes of the white-browed piculet (Sasia ochracea, Picidae) and pale-billed woodpecker (Campephilus guatemalensis, Picidae). Mitochondrial DNA A. 27:3640–3641.
- Gibb GC, Kardailsky O, Kimball RT, Braun EL, Penny D. 2007. Mitochondrial genomes and avian phylogeny: complex characters and resolvability without explosive radiations. Mol Biol Evol. 24:269–280.
- Koh H, Park GS, Shin SM, Park CE, Kim S, Han SJ, Pham HQ, Shin J-H, Lee DW. 2018. Mitochondrial mutations in cholestatic liver disease with biliary atresia. Sci Rep. 8:905.
- Park YJ, Park CE, Lee SH, Ko HS, Ullah I, Hwang UW, Shin JH. 2018. The complete mitochondrial genome sequence of the intertidal crab Parasesarma Tripectinis (Arthropoda, Decapoda, Sesarmidae). Mitochondrial DNA B. 3:193–194.
- Zhang Z, An M, Deng Y, Zhu S. 2016. The complete mitochondrial genome of the Downy woodpecker, Picoides pubescens (Piciformes: Picidae). Mitochondrial DNA A. 27:3479–3480.