Abstract
Euchiloglanis davidi, an endemic species to China, is mainly distributed in the upper Yangtze River and its tributaries. In this study, the complete mitochondrial genome of E. davidi was first sequenced. The result showed that the circular mitogenome was 16,559 bp in length and contained 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, one origin of replication on the light-strand, and one control region. These genes were in the same order as in other vertebrate species. Except ND6 and eight tRNA genes, most mitochondrial genes were encoded on the H-strand. The whole mitogenome base composition of 31.18% A, 25.12% T, 28.09% C, 15.61% G, with 56.30% AT. Phylogenetic analyses using concatenated nucleotide sequences of the 13 protein-coding genes with two different methods (Maximum likelihood and neighbor-joining analysis) both highly supported that E. davidi showed a close relationship with E. kishinouyei. This work provides basic genetic data of E. davidi which will be helpful for further researches on conservation biology and species identification.
Euchiloglanis davidi, belonging to the genus Euchiloglanis within the family Sisoridae, which is an endemic species to China, mainly distributed in upper reaches of the Yangtze River drainage (Ding Citation1994). Recent research has indicated that the natural population number of this fish strongly decreased because of overfishing and habitat degradation (Pan et al. Citation2009). Since the morphological similarities, this species was often misidentified and recorded as other Euchiloglanis during the international trade. Therefore, it is necessary to develop the genetic identification methods of E. davidi, which would be one of the essential approaches to distinguish E. davidi with other Euchiloglanis species. In the present study, we determined the complete mitochondrial genome of E. davidi and constructed the phylogenetic relationship among the Sisoridae species, which expects for the phylogenetic relationships of these taxa and further conservation strategies for the economically important species.
The complete mitochondrial DNA was first determined using the next generation sequencing (NGS). The specimens were obtained from Jintang (30°29′10″N, 104°20′37″E), Chengdu in August 2018, and were stored in Zoological Specimen Museum of Neijiang Normal University (accession number: B0801201807250003). A 30–40 mg fin clip was collected and preserved in 95% ethanol at 4 °C. Total genomic DNA was extracted with a Tissue DNA Kit (OMEGA E.Z.N.A.) following the manufacturer’s protocol. Subsequently, the genomic DNA was sequenced using the NGS, and then the mitogenome was assembled using E. kishinouyei as reference.
The complete assembled mitogenome of E. davidi was a circular molecule with 16,559 bp long (GenBank Accession number MK181572). It was made up of 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and two non-coding regions: origin of light-strand replication (OL) and displacement loop locus (D-loop), which was found to be similar to that of most other vertebrates (Ma et al. Citation2014; Wu et al. Citation2018). The overall nucleotides composition of E. davidi was 31.18% A, 25.12% T, 28.09% C, 15.61% G, with a slight AT bias of 56.30%. There were 37 genes, nine of these genes were encoded on the L-strand, including ND6, tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNAGlu, tRNAPro, tRNATyr, and tRNASer, the remaining 28 genes were encoded on H-strand. In 13 PCGs, the longest was ND5 (1776 bp in length), and the shortest was ATP8 (172 bp in length). Except for ND1 (started with ATA), COX1 (started with GTG), and ND5 (started with GTA), the rest of the PCGs started with ATG. Besides, five PCGs stopped at the incomplete codon (T–), except five genes (ND1, COX2, ND4L, ND5, Cyt b) terminated with TAA codon, two genes (ND2, ND6) used TAG as stop codon and ATP6 stopped at the (TA-). The 12S rRNA(952 bp) and 16S rRNA (1722 bp) were located between tRNALeu and tRNAPhe and separated by tRNAVal gene. There were seven overlapping reading frames in mitogenome of E. davidi, which were respectively 16S rRNA-tRNALeu, tRNAIle-tRNAGln, ND2-tRNATrp, ATP8-ATP6, ND4L-ND4, ND5-ND6, tRNAThr-tRNAPro. There were 22 genes interspersed in rRNAs and PCGs, the lengths range from 67 bp (tRNACys) to 76 bp (tRNAThr). The D-loop region (891 bp) was located between tRNAPro and tRNAPhe.
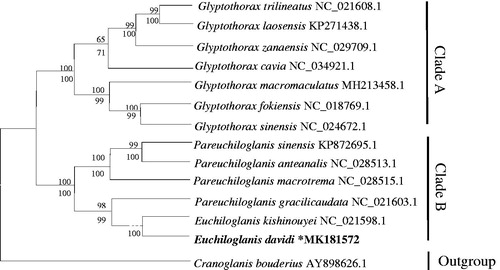
Thus far, the mitochondrial genes have been widely used for inferring phylogenetic relationships (Boore et al. Citation2005). A phylogenetic tree was constructed based on the 13 PCGs nucleotide sequences of E. davidi and other Sisoridae fishes using the neighbor-joining (NJ) method and maximum likelihood (ML) method (Zou et al. Citation2017, Citation2018). The General Time Reversible model and Gamma distribution with Invariant sites (GTR + G+I) were selected as the best phylogenetic model. Among them, Cranoglanis bouderius defined as an alien species. Combined NJ tree and ML tree, the same topological structure tree was obtained. Both trees showed that Sisoridae was divided into two monophyletic clades (A, B) (). The detailed branch situation was as follows: G. trilineatus, G. laosensis, G. zanaensis, G. cavia, G. macromaculatus, G. fokiensis, and G. sinensis were grouped into clade A, and the rest species belonged to clade B. This result highly supported the closest relationship between our sample and E. kishinouyei. Furthermore, this mitochondrial genome (E. davidi) will provide additional insight into evolutionary studies and conservation genetics of this threatened species.
Figure 1. Based on the nucleotide arrangement of 13 protein-coding genes, the phylogenetic relationships among the 13 taxonomic units were indicated by the phylogenetic development of methods maximum likelihood (ML) and neighbour-joining (NJ). The topology tree obtained by ML and NJ analysis is equivalent. The node displays the NJ posterior probability and ML boot value. Cranoglanis bouderius (GenBank:AY898626.1) was used as the outgroup.
Disclosure statement
The authors declare no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Boore JL, Macey JR, Medina M. 2005. Sequencing and comparing whole mitochondrial genomes of animals. Methods Enzymol. 19:311–348.
- Ding RH. (1994). The fishes of Sichuan, China. Chengdu: Sichuan Publishing House of Science and Technology; p. 215–220.
- Ma X, Huang F, Wang Z. 2014. The complete mitochondrial genome sequence of Acrossocheilus monticolus (Teleostei, Cypriniformes, Cyprinidae). Mitochondrial DNA. 25:245–246.
- Pan YY, Feng J, Du WP, Jiang B G, Luo B, Pang W. 2009. The study about rate of flesh in body and nutritional composition of muscle in Euchiloglanic spp. Acta Hydrobiol Sinica. 33:980–985.
- Wu T, Zhang T, Zou YC, Chen M, Deng Q. 2018. The complete mitochondrial genome of Sarcocheilichthys davidi and its phylogeny, Mitochondrial DNA B. 3:412–413.
- Zou YC, Xie BW, Qin CJ, Wang YM, Yuan DY, Li R, Weng ZY. 2017. The complete mitochondrial genome of a threatened loach (Sinibotia reevesae) and its phylogeny. Genes Genom. 39:767–778.
- Zou YC, Zhang JY, Wen ZY, Qin CJ, Wang YM, Li R, Qi ZM. 2018. The complete mitochondrial genome of the hybrid of Acipenser dabryanus (♀) × A. schrenckii (♂) and its phylogeny. Conserv Genet Resour. 10:881–884.
