Abstract
Epipremnum aureum is known as devil’s ivy as it stays green even when kept in the dark. It is an important foliage plant in the Araceae family. In this study, we reported and characterized the complete chloroplast genome sequence of E. aureum 'Neon', a variety that has solid yellow-green leaves with no variegation, obtained using next-generation sequencing. The chloroplast genome was determined to be 163,961 bp in length. It contained large single-copy (LSC) and small single copy (SSC) regions of 89,728 and 22,259 bp, respectively, which were separated by a pair of 25,987 bp inverted repeat (IR) regions. The genome is predicted to contain 122 genes, including 77 protein-coding genes, 37 tRNA genes, and eight rRNA genes. The overall GC content of the genome is 36.1%. A phylogenetic tree reconstructed by 20 chloroplast genomes reveals that E. aureum ‘Neon’ is most closely related to Zantedeschia aethiopica in Araceae.
Epipremnum aureum, commonly known as ‘Golden pothos’, is native to South East Asia and Solomon islands (Huxley et al. Citation1994). It is also known as devil’s ivy as it stays green even when kept in the dark. Epipremnum aureum consists of five varieties, including E. aureum ‘Golden Pothos’, E. aureum ‘Neon’, E. aureum ‘Marble Queen’, E. aureum ‘Jade Pothos’, and E. aureum ‘N Joy’. The kinship between these variants is not very clear due to the lack of genomic information. Here we characterized the complete chloroplast genome sequence of E. aureum ‘Neon’, which has solid yellow-green leaves with no variegation, based on the genome skimming sequencing data. This genome data can provide intragenic information for clarifying the taxonomic identities and valuable information about the evolution of Epipremnum.
The total genomic DNA was extracted from the fresh leaves of E. aureum ‘Neon’(Horticultural Greenhouse in Fujian Agriculture and Forestry University, 26.08°N, 119.23°E) using the DNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA). The DNA was is stored at −80 °C in our lab. The whole genome sequencing was conducted by Nanjing Genepioneer Biotechnologies Inc. (Nanjing, China) on the Illumina Hiseq platform. The filtered sequences were assembled using the program SPAdes assembler 3.10.0 (Bankevich et al. Citation2012). Annotation was performed using the DOGMA (Wyman et al. Citation2004) and BLAST searches. The plastome of E. aureum ‘Neon’ was determined to comprise a double-stranded, circular DNA of 163,961 bp (NCBI acc. no. MK286107), and it contained two inverted repeat (IR) regions of 25,987 bp each, separated by large single-copy (LSC) and small single copy (SSC) regions of 89,728 and 22,259 bp, respectively. The genome was predicted to contain 122 genes, including 77 protein-coding genes, 37 tRNA genes, and eight rRNA genes. Six full protein-coding genes, seven tRNA genes and four rRNA genes were duplicated in IR regions. Sixteen genes contained one intron and two genes (clpP and ycf3) contained two introns. The overall GC content of E. aureum ‘Neon’ cp genome is 36.1% and the corresponding values in LSC, SSC and IR regions are 34.6, 28.6 and 41.9%, respectively.
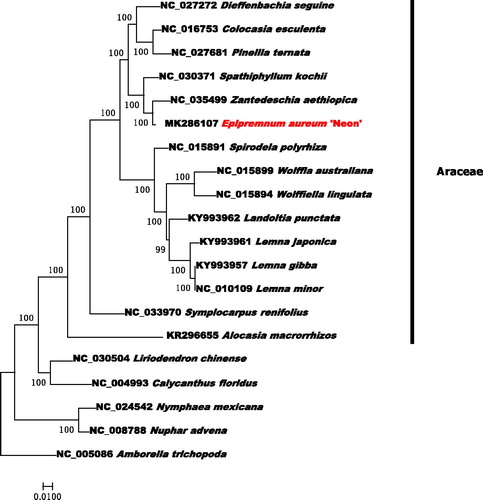
To investigate its taxonomic status, Alignment was performed on the 20 chloroplast genome sequences (Amborella trichopoda was used as an outgroup) using MAFFT v7.307 (Katoh and Standley Citation2013), and a maximum likelihood (ML) tree was constructed by FastTree version 2.1.10 (Price Citation2010). The ML phylogenetic tree shows that E. aureum ‘Neon’ is most closely related to Zantedeschia aethiopica in Araceae, with bootstrap support values of 100% ().
Acknowledgements
The authors are grateful to Mingzhi Li at Genepioneer Biotechnologies Co. Ltd., Nanjing for providing technical assistance.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Pyshkin AV. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Huxley A, Griffiths M, Levy M. Grifiths M: Index of garden plants: The New Horticulture Society Dictionary of Gardening. 4th ed. Vol 1. London: Macmillan; 1994. p 3353.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PloS one. 5(3):e9490.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.